A steam-based method to investigate biofilm
- PMID: 30158585
- PMCID: PMC6115380
- DOI: 10.1038/s41598-018-31437-y
A steam-based method to investigate biofilm
Abstract
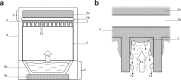
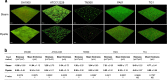
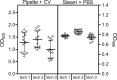
Biofilm has become a major topic of interest in medical, food, industrial, and environmental bacteriology. To be relevant, investigation of biofilm behavior requires effective and reliable techniques. We present herein a simple and robust method, adapted from the microplate technique, in which steam is used as a soft washing method to preserve biofilm integrity and to improve reproducibility of biofilm quantification. The kinetics of steam washing indicated that the method is adapted to remove both planktonic bacteria and excess crystal violet (CV) staining for S. aureus, S. epidermidis, S. carnosus, P. aeruginosa, and E. coli biofilm. Confocal laser scanning microscopy confirmed that steam washing preserved the integrity of the biofilm better than pipette-based washing. We also investigated the measurement of the turbidity of biofilm resuspended in phosphate-buffered saline (PBS) as an alternative to staining with CV. This approach allows the discrimination of biofilm producer strains from non-biofilm producer strains in a way similar to CV staining, and subsequently permits quantification of viable bacteria present in biofilm by culture enumeration from the same well. Biofilm quantification using steam washing and PBS turbidity reduced the technical time needed, and data were highly reproducible.
Conflict of interest statement
The steam washing process was patented (patent applicant: UCBL (Lyon 1), INSERM, CNRS, ENS-Lyon, HCL; inventors: Régis VILLET and Frédéric LAURENT; application number: FR1659459 and PCT/FR2017052646, status: pending).
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

