Emergence of Intrahepatic Cholangiocarcinoma: How High-Throughput Technologies Expedite the Solutions for a Rare Cancer Type
- PMID: 30158952
- PMCID: PMC6104394
- DOI: 10.3389/fgene.2018.00309
Emergence of Intrahepatic Cholangiocarcinoma: How High-Throughput Technologies Expedite the Solutions for a Rare Cancer Type
Abstract
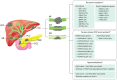
Intrahepatic cholangiocarcinoma (ICC) is the cancer of the intrahepatic bile ducts, and together with hepatocellular carcinoma (HCC), constitute the majority of primary liver cancers. ICC is a rare disorder as its overall incidence is < 1/100,000 in the United States and Europe. However, it shows much higher incidence in particular geographical regions, such as northeastern Thailand, where liver fluke infection is the most common risk factor of ICC. Since the early stages of ICC are often asymptomatic, the patients are usually diagnosed at advanced stages with no effective treatments available, leading to the high mortality rate. In addition, unclear genetic mechanisms, heterogeneous nature, and various etiologies complicate the development of new efficient treatments. Recently, a number of studies have employed high-throughput approaches, including next-generation sequencing and mass spectrometry, in order to understand ICC in different biological aspects. In general, the majority of recurrent genetic alterations identified in ICC are enriched in known tumor suppressor genes and oncogenes, such as mutations in TP53, KRAS, BAP1, ARID1A, IDH1, IDH2, and novel FGFR2 fusion genes. Yet, there are no major driver genes with immediate clinical solutions characterized. Interestingly, recent studies utilized multi-omics data to classify ICC into two main subgroups, one with immune response genes as the main driving factor, while another is enriched with driver mutations in the genes associated with epigenetic regulations, such as IDH1 and IDH2. The two subgroups also show different hypermethylation patterns in the promoter regions. Additionally, the immune response induced by host-pathogen interactions, i.e., liver fluke infection, may further stimulate tumor growth through alterations of the tumor microenvironment. For in-depth functional studies, although many ICC cell lines have been globally established, these homogeneous cell lines may not fully explain the highly heterogeneous genetic contents of this disorder. Therefore, the advent of patient-derived xenograft and 3D patient-derived organoids as new disease models together with the understanding of evolution and genetic alterations of tumor cells at the single-cell resolution will likely become the main focus to fill the current translational research gaps of ICC in the future.
Keywords: disease model; high-throughput technology; integrative multi-omics analysis; intrahepatic cholangiocarcinoma; molecular biomarker; precision oncology; translational medicine.
Figures

References
-
- American Cancer Society, Inc. (2018). Data from: Bile Duct Cancer; Early Detection, Diagnosis, and Staging; Survival Rates for Bile Duct Cancer. Available online at: https://www.cancer.org/content/cancer/en/cancer/bile-duct-cancer/detecti...
-
- Andersen J. B., Spee B., Blechacz B. R., Avital I., Komuta M., Barbour A., et al. . (2013). Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology 142, 1021.e1015–1031.e1015. 10.1053/j.gastro.2011.12.005 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

