Characterisation of porcine epidemic diarrhea virus isolates during the 2014-2015 outbreak in the Philippines
- PMID: 30159369
- PMCID: PMC6111962
- DOI: 10.1007/s13337-018-0470-4
Characterisation of porcine epidemic diarrhea virus isolates during the 2014-2015 outbreak in the Philippines
Abstract
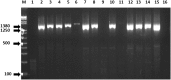
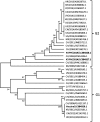
The viral agent of the porcine epidemic diarrhea (PED) was investigated during the reported 2014-2015 outbreaks in commercial farms in Central Luzon, Philippines. The study covered detection of PED virus (PEDV) in fecal and intestinal samples through reverse transcription PCR and sequence analysis of the nucleocapsid (N) gene. Results showed that 10 out of 34 fecal and intestinal samples examined were positive for PEDV. The partial nucleotide sequence of the N gene of the field samples showed 98-99% homologous to PEDV sequences registered in the GenBank. It was also noted that N gene sequences between field samples were 98% homologous. Interestingly, the partial sequences of the N genes of the field samples were genetically similar to the PEDV isolates from USA, China, Mexico, Canada and Japan. The phylogenetic tree analysis revealed that the Philippine samples clustered in group 2-1 of the PEDV, wherein the isolates of this group were responsible for the outbreaks in Asia and the USA. Analysis of the partial nucleotide and amino acid sequences revealed polymorphisms, deletions and insertions in the N-gene of the PEDV. Amino acid sequence alignment also showed deletions and insertion in the PEDV detected in the Philippines.
Keywords: Nucleocapsid gene; Nucleotide polymorphisms; Phylogenetic tree analysis; Porcine epidemic diarrhea virus.
Figures
Similar articles
-
Epidemic strain YC2014 of porcine epidemic diarrhea virus could provide piglets against homologous challenge.Virol J. 2016 Apr 22;13:68. doi: 10.1186/s12985-016-0529-z. Virol J. 2016. PMID: 27103490 Free PMC article.
-
Detection and phylogenetic analysis of porcine epidemic diarrhea virus in central China based on the ORF3 gene and the S1 gene.Virol J. 2016 Nov 25;13(1):192. doi: 10.1186/s12985-016-0646-8. Virol J. 2016. PMID: 27887624 Free PMC article.
-
US-like isolates of porcine epidemic diarrhea virus from Japanese outbreaks between 2013 and 2014.Springerplus. 2015 Dec 2;4:756. doi: 10.1186/s40064-015-1552-z. eCollection 2015. Springerplus. 2015. PMID: 26693114 Free PMC article.
-
Analysis of the spike, ORF3, and nucleocapsid genes of porcine epidemic diarrhea virus circulating on Thai swine farms, 2011-2016.PeerJ. 2019 Apr 30;7:e6843. doi: 10.7717/peerj.6843. eCollection 2019. PeerJ. 2019. PMID: 31106060 Free PMC article.
-
Detection and phylogenetic analyses of spike genes in porcine epidemic diarrhea virus strains circulating in China in 2016-2017.Virol J. 2017 Oct 10;14(1):194. doi: 10.1186/s12985-017-0860-z. Virol J. 2017. PMID: 29017599 Free PMC article.
Cited by
-
Coronaviruses in farm animals: Epidemiology and public health implications.Vet Med Sci. 2021 Mar;7(2):322-347. doi: 10.1002/vms3.359. Epub 2020 Sep 25. Vet Med Sci. 2021. PMID: 32976707 Free PMC article. Review.
-
Advances in porcine epidemic diarrhea virus research: genome, epidemiology, vaccines, and detection methods.Discov Nano. 2025 Mar 3;20(1):48. doi: 10.1186/s11671-025-04220-y. Discov Nano. 2025. PMID: 40029472 Free PMC article. Review.
-
Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in southern China from 2012 to 2018.BMC Vet Res. 2019 Dec 27;15(1):470. doi: 10.1186/s12917-019-2212-2. BMC Vet Res. 2019. PMID: 31881873 Free PMC article.
-
Swine coronaviruses (SCoVs) and their emerging threats to swine population, inter-species transmission, exploring the susceptibility of pigs for SARS-CoV-2 and zoonotic concerns.Vet Q. 2022 Dec;42(1):125-147. doi: 10.1080/01652176.2022.2079756. Vet Q. 2022. PMID: 35584308 Free PMC article. Review.
References
-
- Song DS, Oh JS, Kang BK, Yang JS, Song JY, Moon HJ, Kim TY, Yoo HS, Jang YS, Park BK. Fecal shedding of a highly cell-culture-adapted porcine epidemic virus after oral inoculation in pigs. J Swine Health Prod. 2005;13(5):269–272.
LinkOut - more resources
Full Text Sources
Other Literature Sources