Molecular functions and specific roles of circRNAs in the cardiovascular system
- PMID: 30159442
- PMCID: PMC6096412
- DOI: 10.1016/j.ncrna.2018.05.002
Molecular functions and specific roles of circRNAs in the cardiovascular system
Erratum in
-
Erratum regarding previously published articles.Noncoding RNA Res. 2020 Nov 7;5(4):220-221. doi: 10.1016/j.ncrna.2020.11.002. eCollection 2020 Dec. Noncoding RNA Res. 2020. PMID: 33294748 Free PMC article.
Abstract
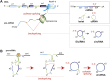
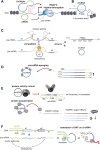
As part of the superfamily of long noncoding RNAs, circular RNAs (circRNAs) are emerging as a new type of regulatory molecules that partake in gene expression control. Here, we review the current knowledge about circRNAs in cardiovascular disease. CircRNAs are not only associated with different types of cardiovascular disease, but they have also been identified as intracellular effector molecules for pathophysiological changes in cardiovascular tissues, and as cardiovascular biomarkers. This evidence is put in the context of the current understanding of general circRNA biogenesis and of known interactions of circRNAs with DNA, RNA, and proteins.
Keywords: Cardiovascular disease; Circular RNA; Non-coding RNA; Splicing; Transcription; circRNA.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

