PRDM9, a driver of the genetic map
- PMID: 30161134
- PMCID: PMC6116924
- DOI: 10.1371/journal.pgen.1007479
PRDM9, a driver of the genetic map
Abstract
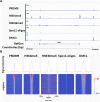
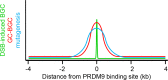
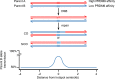
During meiosis, maternal and paternal chromosomes undergo exchanges by homologous recombination. This is essential for fertility and contributes to genome evolution. In many eukaryotes, sites of meiotic recombination, also called hotspots, are regions of accessible chromatin, but in many vertebrates, their location follows a distinct pattern and is specified by PR domain-containing protein 9 (PRDM9). The specification of meiotic recombination hotspots is achieved by the different activities of PRDM9: DNA binding, histone methyltransferase, and interaction with other proteins. Remarkably, PRDM9 activity leads to the erosion of its own binding sites and the rapid evolution of its DNA-binding domain. PRDM9 may also contribute to reproductive isolation, as it is involved in hybrid sterility potentially due to a reduction of its activity in specific heterozygous contexts.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

