Helena and BS: Two Travellers between the Genera Drosophila and Zaprionus
- PMID: 30165545
- PMCID: PMC6179348
- DOI: 10.1093/gbe/evy184
Helena and BS: Two Travellers between the Genera Drosophila and Zaprionus
Abstract
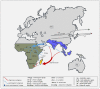
The frequency of horizontal transfers of transposable elements (HTTs) varies among the types of elements according to the transposition mode and the geographical and temporal overlap of the species involved in the transfer. The drosophilid species of the genus Zaprionus and those of the melanogaster, obscura, repleta, and virilis groups of the genus Drosophila investigated in this study shared space and time at some point in their evolutionary history. This is particularly true of the subgenus Zaprionus and the melanogaster subgroup, which overlapped both geographically and temporally in Tropical Africa during their period of origin and diversification. Here, we tested the hypothesis that this overlap may have facilitated the transfer of retrotransposons without long terminal repeats (non-LTRs) between these species. We estimated the HTT frequency of the non-LTRs BS and Helena at the genome-wide scale by using a phylogenetic framework and a vertical and horizontal inheritance consistence analysis (VHICA). An excessively low synonymous divergence among distantly related species and incongruities between the transposable element and species phylogenies allowed us to propose at least four relatively recent HTT events of Helena and BS involving ancestors of the subgroup melanogaster and ancestors of the subgenus Zaprionus during their concomitant diversification in Tropical Africa, along with older possible events between species of the subgenera Drosophila and Sophophora. This study provides the first evidence for HTT of non-LTRs retrotransposons between Drosophila and Zaprionus, including an in-depth reconstruction of the time frame and geography of these events.
Figures



References
-
- Altschul SF, et al. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. - PubMed
-
- Anxolabéhère D, Kidwell MG, Periquet G.. 1988. Molecular characteristics of diverse populations are consistent with the hypothesis of a recent invasion of Drosophila melanogaster by mobile P elements. Mol Biol Evol. 5(3):252–269. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

