Large-scale comparative analysis of microbial pan-genomes using PanOCT
- PMID: 30165579
- PMCID: PMC6419995
- DOI: 10.1093/bioinformatics/bty744
Large-scale comparative analysis of microbial pan-genomes using PanOCT
Abstract
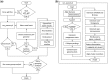
Summary: The JCVI pan-genome pipeline is a collection of programs to run PanOCT and tools that support and extend the capabilities of PanOCT. PanOCT (pan-genome ortholog clustering tool) is a tool for pan-genome analysis of closely related prokaryotic species or strains. The JCVI Pan-Genome Pipeline wrapper invokes command-line utilities that prepare input genomes, invoke third-party tools such as NCBI Blast+, run PanOCT, generate a consensus pan-genome, annotate features of the pan-genome, detect sets of genes of interest such as antimicrobial resistance (AMR) genes and generate figures, tables and html pages to visualize the results. The pipeline can run in a hierarchical mode, lowering the RAM and compute resources used.
Availability and implementation: Source code, demo data, and detailed documentation are freely available at https://github.com/JCVenterInstitute/PanGenomePipeline.
© The Author(s) 2018. Published by Oxford University Press.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials