High-resolution analysis of the pneumococcal transcriptome under a wide range of infection-relevant conditions
- PMID: 30165663
- PMCID: PMC6212715
- DOI: 10.1093/nar/gky750
High-resolution analysis of the pneumococcal transcriptome under a wide range of infection-relevant conditions
Abstract
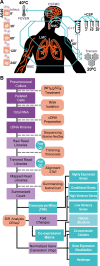
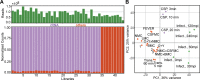
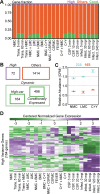
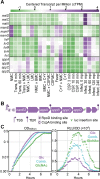
Streptococcus pneumoniae is an opportunistic human pathogen that typically colonizes the nasopharyngeal passage and causes lethal disease in other host niches, such as the lung or the meninges. The expression and regulation of pneumococcal genes at different life-cycle stages, such as commensal or pathogenic, are not entirely understood. To chart the transcriptional responses of S. pneumoniae, we used RNA-seq to quantify the relative abundance of the transcriptome under 22 different infection-relevant conditions. The data demonstrated a high level of dynamic expression and, strikingly, all annotated pneumococcal genomic features were expressed in at least one of the studied conditions. By computing the correlation values of every pair of genes across all studied conditions, we created a co-expression matrix that provides valuable information on both operon structure and regulatory processes. The co-expression data are highly consistent with well-characterized operons and regulons, such as the PyrR, ComE and ComX regulons, and have allowed us to identify a new member of the competence regulon. Lastly, we created an interactive data center named PneumoExpress (https://veeninglab.com/pneumoexpress) that enables users to access the expression data as well as the co-expression matrix in an intuitive and efficient manner, providing a valuable resource to the pneumococcal research community.
Figures




References
-
- Troeger C., Forouzanfar M., Rao P.C., Khalil I., Brown A., Swartz S., Fullman N., Mosser J., Thompson R.L., Reiner R.C. et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. Infect. Dis. 2017; 17:1133–1161. - PMC - PubMed
-
- Kassebaum N.J., Arora M., Barber R.M., Bhutta Z.A., Brown J., Carter A., Casey D.C., Charlson F.J., Coates M.M., Coggeshall M. et al. Global, regional, and national disability-adjusted life-years (DALYs) for 315 diseases and injuries and healthy life expectancy (HALE), 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016; 388:1603–1658. - PMC - PubMed
-
- Wardlaw T., Salama P., Johansson E.W., Mason E.. Pneumonia: the leading killer of children. Lancet. 2006; 368:1048–1050. - PubMed
-
- Welte T., Torres A., Nathwani D.. Clinical and economic burden of community-acquired pneumonia among adults in Europe. Thorax. 2012; 67:71–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

