Selection of marker genes for genetic barcoding of microorganisms and binning of metagenomic reads by Barcoder software tools
- PMID: 30165813
- PMCID: PMC6117900
- DOI: 10.1186/s12859-018-2320-1
Selection of marker genes for genetic barcoding of microorganisms and binning of metagenomic reads by Barcoder software tools
Abstract
Background: Metagenomic approaches have revealed the complexity of environmental microbiomes with the advancement in whole genome sequencing displaying a significant level of genetic heterogeneity on the species level. It has become apparent that patterns of superior bioactivity of bacteria applicable in biotechnology as well as the enhanced virulence of pathogens often requires distinguishing between closely related species or sub-species. Current methods for binning of metagenomic reads usually do not allow for identification below the genus level and generally stops at the family level.
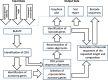
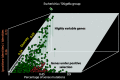
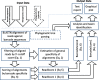
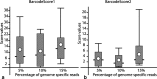
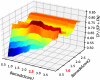
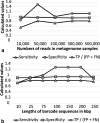
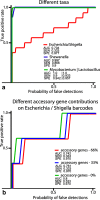
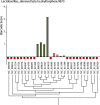
Results: In this work, an attempt was made to improve metagenomic binning resolution by creating genome specific barcodes based on the core and accessory genomes. This protocol was implemented in novel software tools available for use and download from http://bargene.bi.up.ac.za /. The most abundant barcode genes from the core genomes were found to encode for ribosomal proteins, certain central metabolic genes and ABC transporters. Performance of metabarcode sequences created by this package was evaluated using artificially generated and publically available metagenomic datasets. Furthermore, a program (Barcoding 2.0) was developed to align reads against barcode sequences and thereafter calculate various parameters to score the alignments and the individual barcodes. Taxonomic units were identified in metagenomic samples by comparison of the calculated barcode scores to set cut-off values. In this study, it was found that varying sample sizes, i.e. number of reads in a metagenome and metabarcode lengths, had no significant effect on the sensitivity and specificity of the algorithm. Receiver operating characteristics (ROC) curves were calculated for different taxonomic groups based on the results of identification of the corresponding genomes in artificial metagenomic datasets. The reliability of distinguishing between species of the same genus or family by the program was nearly perfect.
Conclusions: The results showed that the novel online tool BarcodeGenerator ( http://bargene.bi.up.ac.za /) is an efficient approach for generating barcode sequences from a set of complete genomes provided by users. Another program, Barcoder 2.0 is available from the same resource to enable an efficient and practical use of metabarcodes for visualization of the distribution of organisms of interest in environmental and clinical samples.
Keywords: Bacterial genome; Metabarcoding; Metagenome; NGS; Software tool.
Conflict of interest statement
Ethics approval and consent to participate
No ethics committee approval is required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Exploiting topic modeling to boost metagenomic reads binning.BMC Bioinformatics. 2015;16 Suppl 5(Suppl 5):S2. doi: 10.1186/1471-2105-16-S5-S2. Epub 2015 Mar 18. BMC Bioinformatics. 2015. PMID: 25859745 Free PMC article.
-
MetaCluster-TA: taxonomic annotation for metagenomic data based on assembly-assisted binning.BMC Genomics. 2014;15 Suppl 1(Suppl 1):S12. doi: 10.1186/1471-2164-15-S1-S12. Epub 2014 Jan 24. BMC Genomics. 2014. PMID: 24564377 Free PMC article.
-
MinION™ nanopore sequencing of environmental metagenomes: a synthetic approach.Gigascience. 2017 Mar 1;6(3):1-10. doi: 10.1093/gigascience/gix007. Gigascience. 2017. PMID: 28327976 Free PMC article.
-
Genome-resolved metagenomics using environmental and clinical samples.Brief Bioinform. 2021 Sep 2;22(5):bbab030. doi: 10.1093/bib/bbab030. Brief Bioinform. 2021. PMID: 33758906 Free PMC article. Review.
-
Fungal DNA barcoding.Genome. 2016 Nov;59(11):913-932. doi: 10.1139/gen-2016-0046. Epub 2016 Aug 30. Genome. 2016. PMID: 27829306 Review.
Cited by
-
Exploring microbial functional biodiversity at the protein family level-From metagenomic sequence reads to annotated protein clusters.Front Bioinform. 2023 Mar 3;3:1157956. doi: 10.3389/fbinf.2023.1157956. eCollection 2023. Front Bioinform. 2023. PMID: 36959975 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

