The CAIRR Pipeline for Submitting Standards-Compliant B and T Cell Receptor Repertoire Sequencing Studies to the National Center for Biotechnology Information Repositories
- PMID: 30166985
- PMCID: PMC6105692
- DOI: 10.3389/fimmu.2018.01877
The CAIRR Pipeline for Submitting Standards-Compliant B and T Cell Receptor Repertoire Sequencing Studies to the National Center for Biotechnology Information Repositories
Abstract
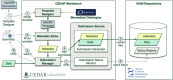
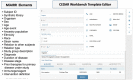
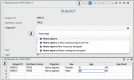
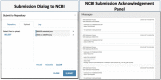
The adaptation of high-throughput sequencing to the B cell receptor and T cell receptor has made it possible to characterize the adaptive immune receptor repertoire (AIRR) at unprecedented depth. These AIRR sequencing (AIRR-seq) studies offer tremendous potential to increase the understanding of adaptive immune responses in vaccinology, infectious disease, autoimmunity, and cancer. The increasingly wide application of AIRR-seq is leading to a critical mass of studies being deposited in the public domain, offering the possibility of novel scientific insights through secondary analyses and meta-analyses. However, effective sharing of these large-scale data remains a challenge. The AIRR community has proposed minimal information about adaptive immune receptor repertoire (MiAIRR), a standard for reporting AIRR-seq studies. The MiAIRR standard has been operationalized using the National Center for Biotechnology Information (NCBI) repositories. Submissions of AIRR-seq data to the NCBI repositories typically use a combination of web-based and flat-file templates and include only a minimal amount of terminology validation. As a result, AIRR-seq studies at the NCBI are often described using inconsistent terminologies, limiting scientists' ability to access, find, interoperate, and reuse the data sets. In order to improve metadata quality and ease submission of AIRR-seq studies to the NCBI, we have leveraged the software framework developed by the Center for Expanded Data Annotation and Retrieval (CEDAR), which develops technologies involving the use of data standards and ontologies to improve metadata quality. The resulting CEDAR-AIRR (CAIRR) pipeline enables data submitters to: (i) create web-based templates whose entries are controlled by ontology terms, (ii) generate and validate metadata, and (iii) submit the ontology-linked metadata and sequence files (FASTQ) to the NCBI BioProject, BioSample, and Sequence Read Archive databases. Overall, CAIRR provides a web-based metadata submission interface that supports compliance with the MiAIRR standard. This pipeline is available at http://cairr.miairr.org, and will facilitate the NCBI submission process and improve the metadata quality of AIRR-seq studies.
Keywords: B cell receptor; National Center for Biotechnology Information; Rep-seq; T cell receptor; antibody; immune-repertoire sequencing; ontology.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

