Crystal structure of the ligand-binding domain of a LysR-type transcriptional regulator: transcriptional activation via a rotary switch
- PMID: 30168204
- PMCID: PMC6886530
- DOI: 10.1111/mmi.14115
Crystal structure of the ligand-binding domain of a LysR-type transcriptional regulator: transcriptional activation via a rotary switch
Abstract
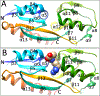
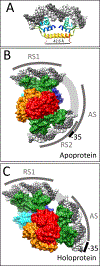
LysR-type transcriptional regulators (LTTRs) generally bind to target promoters in two conformations, depending on the availability of inducing ligands. OccR is an LTTR that regulates the octopine catabolism operon of Agrobacterium tumefaciens. OccR binds to a site located between the divergent occQ and occR promoters. Octopine triggers a conformational change that activates the occQ promoter, and does not affect autorepression. This change shortens the length of bound DNA and relaxes a high-angle DNA bend. Here, we describe the crystal structure of the ligand-binding domain (LBD) of OccR apoprotein and holoprotein. Pairs of LBDs form dimers with extensive hydrogen bonding, while pairs of dimers interact via a single helix, creating a tetramer interface. Octopine causes a 70° rotation of each dimer with respect to the opposite dimer, precisely at the tetramer interface. We modeled the DNA binding domain (DBD), linker helix and bound DNA onto the apoprotein and holoprotein. The two DBDs of the modeled apoprotein lie far apart and the bound DNA between them has a high-angle DNA bend. In contrast, the two DBDs of the holoprotein lie closer to each other, with a low DNA bend angle. This inter-dimer pivot fully explains earlier studies of this LTTR.
© 2018 John Wiley & Sons Ltd.
Figures





Similar articles
-
Most mutant OccR proteins that are defective in positive control hold operator DNA in a locked high-angle bend.J Bacteriol. 2011 Oct;193(19):5442-9. doi: 10.1128/JB.05352-11. Epub 2011 Jul 29. J Bacteriol. 2011. PMID: 21804007 Free PMC article.
-
Altered-function mutations in the Agrobacterium tumefaciens OccR protein and in an OccR-regulated promoter.J Bacteriol. 1993 Dec;175(23):7715-9. doi: 10.1128/jb.175.23.7715-7719.1993. J Bacteriol. 1993. PMID: 8244944 Free PMC article.
-
Mutations in the occQ operator that decrease OccR-induced DNA bending do not cause constitutive promoter activity.J Biol Chem. 2002 May 3;277(18):15773-80. doi: 10.1074/jbc.M200109200. Epub 2002 Feb 27. J Biol Chem. 2002. PMID: 11877409
-
Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins.Microbiology (Reading). 2008 Dec;154(Pt 12):3609-3623. doi: 10.1099/mic.0.2008/022772-0. Microbiology (Reading). 2008. PMID: 19047729 Review.
-
Molecular biology of the LysR family of transcriptional regulators.Annu Rev Microbiol. 1993;47:597-626. doi: 10.1146/annurev.mi.47.100193.003121. Annu Rev Microbiol. 1993. PMID: 8257110 Review.
Cited by
-
The LysR-Type Transcriptional Regulator YeeY Plays Important Roles in the Regulatory of Furazolidone Resistance in Aeromonas hydrophila.Front Microbiol. 2020 Sep 9;11:577376. doi: 10.3389/fmicb.2020.577376. eCollection 2020. Front Microbiol. 2020. PMID: 33013815 Free PMC article.
-
Transcriptional regulation of two redundant 3-bromo-4-hydroxybenzoate catabolic operons via two different regulatory modes in Pigmentiphaga kullae strain H8.Appl Environ Microbiol. 2025 Apr 23;91(4):e0240324. doi: 10.1128/aem.02403-24. Epub 2025 Mar 4. Appl Environ Microbiol. 2025. PMID: 40035602 Free PMC article.
-
Characterization of the pleiotropic LysR-type transcription regulator LeuO of Escherichia coli.Nucleic Acids Res. 2019 Aug 22;47(14):7363-7379. doi: 10.1093/nar/gkz506. Nucleic Acids Res. 2019. PMID: 31184713 Free PMC article.
-
Mutation of smeRv Renders Stenotrophomonas maltophilia Resistant to First-Line Antibiotics Trimethoprim/Sulfamethoxazole and Levofloxacin.Antibiotics (Basel). 2025 May 28;14(6):550. doi: 10.3390/antibiotics14060550. Antibiotics (Basel). 2025. PMID: 40558140 Free PMC article.
-
Structural dynamics in the evolution of a bilobed protein scaffold.Proc Natl Acad Sci U S A. 2021 Dec 7;118(49):e2026165118. doi: 10.1073/pnas.2026165118. Proc Natl Acad Sci U S A. 2021. PMID: 34845009 Free PMC article.
References
-
- Akakura R, and Winans SC (2002a) Mutations in the occQ operator that decrease OccR-induced DNA bending do not cause constitutive promoter activity. J. Biol. Chem. 277: 15773–15780. - PubMed
-
- Akakura R, and Winans SC (2002b) Constitutive mutations of the OccR regulatory protein affect DNA bending in response to metabolites released from plant tumors. J. Biol. Chem. 277: 5866–5874. - PubMed
-
- Alanazi AM, Neidle EL, and Momany C (2013) The DNA-binding domain of BenM reveals the structural basis for the recognition of a T-N11-A sequence motif by LysR-type transcriptional regulators. Acta. Crystallogr. D Biol. Crystallogr. 69: 1995–2007. - PubMed
-
- Bourras S, Rouxel T, and Meyer M (2015) Agrobacterium tumefaciens gene transfer: how a plant pathogen hacks the nuclei of plant and nonplant organisms. Phytopathology 105: 1288–1301. - PubMed
-
- Brown NL, Stoyanov JV, Kidd SP, and Hobman JL (2003) The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27: 145–163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

