Hotspot mutations and ColE1 plasmids contribute to the fitness of Salmonella Heidelberg in poultry litter
- PMID: 30169497
- PMCID: PMC6118388
- DOI: 10.1371/journal.pone.0202286
Hotspot mutations and ColE1 plasmids contribute to the fitness of Salmonella Heidelberg in poultry litter
Abstract
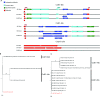
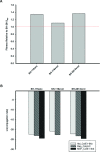
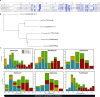
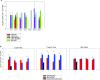
Salmonella enterica subsp. enterica serovar Heidelberg (S. Heidelberg) is a clinically-important serovar linked to food-borne illness, and commonly isolated from poultry. Investigations of a large, multistate outbreak in the USA in 2013 identified poultry litter (PL) as an important extra-intestinal environment that may have selected for specific S. Heidelberg strains. Poultry litter is a mixture of bedding materials and chicken excreta that contains chicken gastrointestinal (GI) bacteria, undigested feed, feathers, and other materials of chicken origin. In this study, we performed a series of controlled laboratory experiments which assessed the microevolution of two S. Heidelberg strains (SH-2813 and SH-116) in PL previously used to raise 3 flocks of broiler chickens. The strains are closely related at the chromosome level, differing from the reference genome by 109 and 89 single nucleotide polymorphisms/InDels, respectively. Whole genome sequencing was performed on 86 isolates recovered after 0, 1, 7 and 14 days of microevolution in PL. Only strains carrying an IncX1 (37kb), 2 ColE1 (4 and 6kb) and 1 ColpVC (2kb) plasmids survived more than 7 days in PL. Competition experiments showed that carriage of these plasmids was associated with increased fitness. This increased fitness was associated with an increased copy number of IncX1 and ColE1 plasmids. Further, all Col plasmid-bearing strains had hotspot mutations in 37 loci on the chromosome and in 3 loci on the IncX1 plasmid. Additionally, we observed a decrease in susceptibility to tobramycin, kanamycin, gentamicin, neomycin and fosfomycin for Col plasmid-bearing strains. Our study demonstrates how positive selection from poultry litter can change the evolutionary path of S. Heidelberg.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Horizontal Gene Transfer and Acquired Antibiotic Resistance in Salmonella enterica Serovar Heidelberg following In Vitro Incubation in Broiler Ceca.Appl Environ Microbiol. 2019 Oct 30;85(22):e01903-19. doi: 10.1128/AEM.01903-19. Print 2019 Nov 15. Appl Environ Microbiol. 2019. PMID: 31471306 Free PMC article.
-
Genomic Comparison of Non-Typhoidal Salmonella enterica Serovars Typhimurium, Enteritidis, Heidelberg, Hadar and Kentucky Isolates from Broiler Chickens.PLoS One. 2015 Jun 17;10(6):e0128773. doi: 10.1371/journal.pone.0128773. eCollection 2015. PLoS One. 2015. PMID: 26083489 Free PMC article.
-
A Whole-Genome Sequencing Approach To Study Cefoxitin-Resistant Salmonella enterica Serovar Heidelberg Isolates from Various Sources.Antimicrob Agents Chemother. 2017 Mar 24;61(4):e01919-16. doi: 10.1128/AAC.01919-16. Print 2017 Apr. Antimicrob Agents Chemother. 2017. PMID: 28137797 Free PMC article.
-
Salmonella Heidelberg isolates from poultry in Brazil and the United States share a large number of resistance and virulence determinants.Microb Pathog. 2025 Jul;204:107523. doi: 10.1016/j.micpath.2025.107523. Epub 2025 Apr 1. Microb Pathog. 2025. PMID: 40180234
-
Recent Evolution and Genomic Profile of Salmonella enterica Serovar Heidelberg Isolates from Poultry Flocks in Brazil.Appl Environ Microbiol. 2021 Oct 14;87(21):e0103621. doi: 10.1128/AEM.01036-21. Epub 2021 Aug 18. Appl Environ Microbiol. 2021. PMID: 34406824 Free PMC article.
Cited by
-
Bacterial Detection and Recovery From Poultry Litter.Front Microbiol. 2022 Jan 6;12:803150. doi: 10.3389/fmicb.2021.803150. eCollection 2021. Front Microbiol. 2022. PMID: 35069507 Free PMC article.
-
Assessment of plasmids for relating the 2020 Salmonella enterica serovar Newport onion outbreak to farms implicated by the outbreak investigation.BMC Genomics. 2023 Apr 4;24(1):165. doi: 10.1186/s12864-023-09245-0. BMC Genomics. 2023. PMID: 37016310 Free PMC article.
-
Iron-Uptake Systems of Chicken-Associated Salmonella Serovars and Their Role in Colonizing the Avian Host.Microorganisms. 2020 Aug 7;8(8):1203. doi: 10.3390/microorganisms8081203. Microorganisms. 2020. PMID: 32784620 Free PMC article. Review.
-
A Familiar Outbreak of Monophasic Salmonella serovar Typhimurium (ST34) Involving Three Dogs and Their Owner's Children.Pathogens. 2022 Dec 8;11(12):1500. doi: 10.3390/pathogens11121500. Pathogens. 2022. PMID: 36558834 Free PMC article.
-
Comprehensive Genomic Characterization of Antibiotic Resistance, Virulence, and Clonality in Salmonella Isolates from Wild Animals in Algeria.Ecohealth. 2023 Dec;20(4):343-348. doi: 10.1007/s10393-023-01670-7. Epub 2024 Jan 4. Ecohealth. 2023. PMID: 38177562
References
-
- USDA, NASS. Poultry production and value—2013 summary. ISSN:1949-1573 ed2014. p. 1–14. Retrieved from “https://usda.mannlib.cornell.edu/usda/nass/PoulProdVa//2010s/2014/PoulPr...”.
-
- MacDonald JM. The economic organization of U.S. broiler production. Washington, D.C.: Economic Research Service, USDA; 2008 June 2008. Report No.: EIB- 38 Contract No.: EIB- 38.
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

