Repertoire of plant RING E3 ubiquitin ligases revisited: New groups counting gene families and single genes
- PMID: 30169501
- PMCID: PMC6118397
- DOI: 10.1371/journal.pone.0203442
Repertoire of plant RING E3 ubiquitin ligases revisited: New groups counting gene families and single genes
Abstract
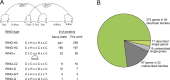
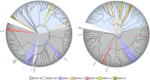
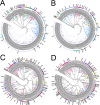
E3 ubiquitin ligases of the ubiquitin proteasome system (UPS) mediate recognition of substrates and later transfer the ubiquitin (Ub). They are the most expanded components of the system. The Really Interesting New Gene (RING) domain contains 40-60 residues that are highly represented among E3 ubiquitin ligases. The Arabidopsis thaliana E3 ubiquitin ligases with a RING finger primarily contain RING-HC or RING-H2 type domains or less frequently RING-v, RING-C2, RING-D, RING-S/T and RING-G type domains. Our previous work on three E3 ubiquitin ligase families with a RING-H2 type domain, ATL, BTL, and CTL, suggested that a phylogenetic distribution based on the RING domain allowed for the creation a catalog of known domains or unknown conserved motifs. This work provided a useful and comprehensive view of particular families of RING E3 ubiquitin ligases. We updated the annotation of A. thaliana RING proteins and surveyed RING proteins from 30 species across eukaryotes. Based on domain architecture profile of the A. thaliana proteins, we catalogued 4711 RING finger proteins into 107 groups, including 66 previously described gene families or single genes and 36 novel families or undescribed genes. Forty-four groups were specific to a plant lineage while 41 groups consisted of proteins found in all eukaryotic species. Our present study updates the current classification of plant RING finger proteins and reiterates the importance of these proteins in plant growth and adaptation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
ATLs and BTLs, plant-specific and general eukaryotic structurally-related E3 ubiquitin ligases.Plant Sci. 2014 Feb;215-216:69-75. doi: 10.1016/j.plantsci.2013.10.017. Epub 2013 Nov 5. Plant Sci. 2014. PMID: 24388516 Review.
-
Expansion and diversification of BTL ring-H2 ubiquitin ligases in angiosperms: putative Rabring7/BCA2 orthologs.PLoS One. 2013 Aug 8;8(8):e72729. doi: 10.1371/journal.pone.0072729. eCollection 2013. PLoS One. 2013. PMID: 23951330 Free PMC article.
-
Diversity in the architecture of ATLs, a family of plant ubiquitin-ligases, leads to recognition and targeting of substrates in different cellular environments.PLoS One. 2011;6(8):e23934. doi: 10.1371/journal.pone.0023934. Epub 2011 Aug 24. PLoS One. 2011. PMID: 21887349 Free PMC article.
-
CTLs, a new class of RING-H2 ubiquitin ligases uncovered by YEELL, a motif close to the RING domain that is present across eukaryotes.PLoS One. 2018 Jan 11;13(1):e0190969. doi: 10.1371/journal.pone.0190969. eCollection 2018. PLoS One. 2018. PMID: 29324855 Free PMC article.
-
Research Progress on Plant RING-Finger Proteins.Genes (Basel). 2019 Nov 26;10(12):973. doi: 10.3390/genes10120973. Genes (Basel). 2019. PMID: 31779262 Free PMC article. Review.
Cited by
-
The ubiquitin-protein ligase MIEL1 localizes to peroxisomes to promote seedling oleosin degradation and lipid droplet mobilization.Proc Natl Acad Sci U S A. 2023 Jul 18;120(29):e2304870120. doi: 10.1073/pnas.2304870120. Epub 2023 Jul 6. Proc Natl Acad Sci U S A. 2023. PMID: 37410814 Free PMC article.
-
Role of Arabidopsis monomeric E3 ubiquitin ligases in the ABA signaling pathway.BMB Rep. 2025 Apr;58(4):147-157. doi: 10.5483/BMBRep.2024-0146. BMB Rep. 2025. PMID: 40058874 Free PMC article. Review.
-
Overview of RING gene family in maize (Zea mays L.): ZmRING-93 enhances drought tolerance in transgenic Arabidopsis.BMC Plant Biol. 2025 May 22;25(1):678. doi: 10.1186/s12870-025-06683-8. BMC Plant Biol. 2025. PMID: 40405093 Free PMC article.
-
Harnessing the ubiquitin code to respond to environmental cues.Essays Biochem. 2022 Aug 5;66(2):111-121. doi: 10.1042/EBC20210094. Essays Biochem. 2022. PMID: 35880291 Free PMC article.
-
The Ubiquitin-Proteasome System (UPS) and Viral Infection in Plants.Plants (Basel). 2022 Sep 22;11(19):2476. doi: 10.3390/plants11192476. Plants (Basel). 2022. PMID: 36235343 Free PMC article. Review.
References
-
- Hershko A, Ciechanover A. The ubiquitin system. Annual Reviews 4139 El Camino Way, PO Box 10139, Palo Alto, CA 94303–0139, USA; 1998.
-
- Aguilar RC, Wendland B. Ubiquitin: not just for proteasomes anymore. Current opinion in cell biology. 2003;15(2):184–90. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

