Widespread evolutionary crosstalk among protein domains in the context of multi-domain proteins
- PMID: 30169546
- PMCID: PMC6118372
- DOI: 10.1371/journal.pone.0203085
Widespread evolutionary crosstalk among protein domains in the context of multi-domain proteins
Abstract
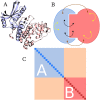
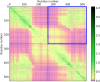
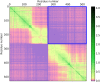
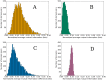
Domains are distinct units within proteins that typically can fold independently into recognizable three-dimensional structures to facilitate their functions. The structural and functional independence of protein domains is reflected by their apparent modularity in the context of multi-domain proteins. In this work, we examined the coupling of evolution of domain sequences co-occurring within multi-domain proteins to see if it proceeds independently, or in a coordinated manner. We used continuous information theory measures to assess the extent of correlated mutations among domains in multi-domain proteins from organisms across the tree of life. In all multi-domain architectures we examined, domains co-occurring within protein sequences had to some degree undergone concerted evolution. This finding challenges the notion of complete modularity and independence of protein domains, providing new perspective on the evolution of protein sequence and function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
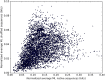
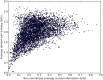
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

