Comparative analysis of methods for evaluation of protein models against native structures
- PMID: 30169622
- PMCID: PMC6419896
- DOI: 10.1093/bioinformatics/bty760
Comparative analysis of methods for evaluation of protein models against native structures
Abstract
Motivation: Measuring discrepancies between protein models and native structures is at the heart of development of protein structure prediction methods and comparison of their performance. A number of different evaluation methods have been developed; however, their comprehensive and unbiased comparison has not been performed.
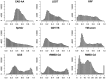
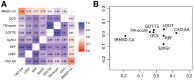
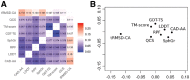
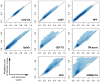
Results: We carried out a comparative analysis of several popular model assessment methods (RMSD, TM-score, GDT, QCS, CAD-score, LDDT, SphereGrinder and RPF) to reveal their relative strengths and weaknesses. The analysis, performed on a large and diverse model set derived in the course of three latest community-wide CASP experiments (CASP10-12), had two major directions. First, we looked at general differences between the scores by analyzing distribution, correspondence and correlation of their values as well as differences in selecting best models. Second, we examined the score differences taking into account various structural properties of models (stereochemistry, hydrogen bonds, packing of domains and chain fragments, missing residues, protein length and secondary structure). Our results provide a solid basis for an informed selection of the most appropriate score or combination of scores depending on the task at hand.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2018. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures







References
-
- Altman N., Krzywinski M. (2015) Association, correlation and causation. Nat. Methods, 12, 899–900. - PubMed
-
- Fisher R.A. (1915) Frequency distribution of the values of the correlation coefficient in samples of an indefinitely large population. Biometrika, 10, 507–521.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

