Early-Life Neuronal-Specific Iron Deficiency Alters the Adult Mouse Hippocampal Transcriptome
- PMID: 30169712
- PMCID: PMC6258792
- DOI: 10.1093/jn/nxy125
Early-Life Neuronal-Specific Iron Deficiency Alters the Adult Mouse Hippocampal Transcriptome
Abstract
Background: Iron deficiency (ID) compromises the developing nervous system, including the hippocampus, resulting in later-life deficits despite iron repletion. The iron-dependent molecular changes driving these lasting deficits, and the effect of early iron repletion, are incompletely understood. Previous studies have utilized dietary models of maternal-fetal ID anemia (IDA) to address these questions; however, concurrent anemia prevents delineation of the specific role of iron.
Objective: The aim of the study was to isolate the effects of developmental ID on adult hippocampal gene expression and to determine if iron repletion reverses these effects in a mouse model of nonanemic hippocampal neuronal ID.
Methods: Nonanemic, hippocampus-specific neuronal ID was generated by using a Tet-OFF dominant negative transferrin receptor (DN-TFR1) mouse model that impairs cellular iron uptake. Hippocampal ID was reversed with doxycycline at postnatal day 21 (P21) in a subset of mice to create 2 experimental groups, chronically iron-deficient and formerly iron-deficient mice, which were compared with their respective doxycycline-treated and untreated iron-sufficient controls. RNA from adult male hippocampi was sequenced. Paired-end reads were analyzed for differential expression. Differentially expressed genes were analyzed in Ingenuity Pathway Analysis.
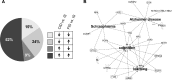
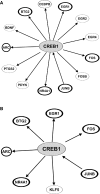
Results: A total of 346 genes were differentially expressed in adult, chronically iron-deficient hippocampi compared with controls. ID dysregulated genes in critical neurodevelopmental pathways, including axonal guidance, CDK5, Ephrin receptor, Rac, and Neurotrophin/Trk signaling. Iron repletion at P21 normalized adult hippocampal expression of 198 genes; however, genes involved in cAMP response element-binding protein (CREB) signaling, neurocognition, and neurologic disease remained dysregulated in adulthood.
Conclusions: Chronic ID during development, independent of anemia, alters the adult mouse hippocampal transcriptome. Restoring iron status during a known critical period of hippocampal neurodevelopment incompletely normalized these changes, suggesting a need for additional studies to identify the most effective timeline for iron therapy, and adjunctive treatments that can fully restore ID-induced molecular changes, particularly in human populations in whom chronic ID is endemic.
Figures
Comment in
-
Examining Consequence of Brain Iron Deficiency in the Absence of Anemia.J Nutr. 2018 Oct 1;148(10):1511-1512. doi: 10.1093/jn/nxy186. J Nutr. 2018. PMID: 30169646 No abstract available.
Similar articles
-
Temporal manipulation of transferrin-receptor-1-dependent iron uptake identifies a sensitive period in mouse hippocampal neurodevelopment.Hippocampus. 2012 Aug;22(8):1691-702. doi: 10.1002/hipo.22004. Epub 2012 Feb 27. Hippocampus. 2012. PMID: 22367974 Free PMC article.
-
Neuronal-specific iron deficiency dysregulates mammalian target of rapamycin signaling during hippocampal development in nonanemic genetic mouse models.J Nutr. 2013 Mar;143(3):260-6. doi: 10.3945/jn.112.168617. Epub 2013 Jan 9. J Nutr. 2013. PMID: 23303869 Free PMC article.
-
Iron Deficiency Impairs Developing Hippocampal Neuron Gene Expression, Energy Metabolism, and Dendrite Complexity.Dev Neurosci. 2016;38(4):264-276. doi: 10.1159/000448514. Epub 2016 Sep 27. Dev Neurosci. 2016. PMID: 27669335 Free PMC article.
-
The role of iron in learning and memory.Adv Nutr. 2011 Mar;2(2):112-21. doi: 10.3945/an.110.000190. Epub 2011 Mar 10. Adv Nutr. 2011. PMID: 22332040 Free PMC article. Review.
-
Iron assessment to protect the developing brain.Am J Clin Nutr. 2017 Dec;106(Suppl 6):1588S-1593S. doi: 10.3945/ajcn.117.155846. Epub 2017 Oct 25. Am J Clin Nutr. 2017. PMID: 29070550 Free PMC article. Review.
Cited by
-
Developmental Iron Deficiency Dysregulates TET Activity and DNA Hydroxymethylation in the Rat Hippocampus and Cerebellum.Dev Neurosci. 2022;44(2):80-90. doi: 10.1159/000521704. Epub 2022 Jan 11. Dev Neurosci. 2022. PMID: 35016180 Free PMC article.
-
Intranasal insulin treatment partially corrects the altered gene expression profile in the hippocampus of developing rats with perinatal iron deficiency.Am J Physiol Regul Integr Comp Physiol. 2023 Oct 1;325(4):R423-R432. doi: 10.1152/ajpregu.00311.2022. Epub 2023 Aug 21. Am J Physiol Regul Integr Comp Physiol. 2023. PMID: 37602386 Free PMC article.
-
Prenatal Iron Deficiency and Choline Supplementation Interact to Epigenetically Regulate Jarid1b and Bdnf in the Rat Hippocampus into Adulthood.Nutrients. 2021 Dec 17;13(12):4527. doi: 10.3390/nu13124527. Nutrients. 2021. PMID: 34960080 Free PMC article.
-
The nAChR Chaperone TMEM35a (NACHO) Contributes to the Development of Hyperalgesia in Mice.Neuroscience. 2021 Mar 1;457:74-87. doi: 10.1016/j.neuroscience.2020.12.027. Epub 2021 Jan 7. Neuroscience. 2021. PMID: 33422618 Free PMC article.
-
The effect of a novel anticonvulsant chemical Q808 on gut microbiota and hippocampus neurotransmitters in pentylenetetrazole-induced seizures in rats.BMC Neurosci. 2022 Feb 3;23(1):7. doi: 10.1186/s12868-022-00690-3. BMC Neurosci. 2022. PMID: 35114941 Free PMC article.
References
-
- Bailey RL, West KP, Black RE. The epidemiology of global micronutrient deficiencies. Ann Nutr Metab 2015;66(Suppl 2):22–33. - PubMed
-
- McLean E, Cogswell M, Egli I, Wojdyla D, de Benoist B. Worldwide prevalence of anaemia: WHO Vitamin and Mineral Nutrition Information System, 1993–2005. Public Health Nutr 2009;12:444–54. - PubMed
-
- Stevens GA, Finucane MM, De-Regil M, Paciorek CJ, Flaxman SR, Branca F, Peña-Rosas JP, Qar Z, Bhutta A, Ezzati M. Global, regional, and national trends in haemoglobin concentration and prevalence of total and severe anaemia in children and pregnant and non-pregnant women for 1995–2011: a systematic analysis of population-representative data. Lancet Glob Health 2013;1:e16–25. - PMC - PubMed
-
- Lozoff B. Iron deficiency and child development. Food Nutr Bull 2007;28:S560–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

