Super-enhancers are transcriptionally more active and cell type-specific than stretch enhancers
- PMID: 30169995
- PMCID: PMC6284781
- DOI: 10.1080/15592294.2018.1514231
Super-enhancers are transcriptionally more active and cell type-specific than stretch enhancers
Abstract
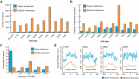
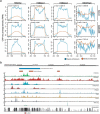
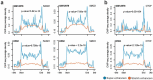
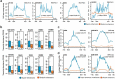
Super-enhancers and stretch enhancers represent classes of transcriptional enhancers that have been shown to control the expression of cell identity genes and carry disease- and trait-associated variants. Specifically, super-enhancers are clusters of enhancers defined based on the binding occupancy of master transcription factors, chromatin regulators, or chromatin marks, while stretch enhancers are large chromatin-defined regulatory regions of at least 3,000 base pairs. Several studies have characterized these regulatory regions in numerous cell types and tissues to decipher their functional importance. However, the differences and similarities between these regulatory regions have not been fully assessed. We integrated genomic, epigenomic, and transcriptomic data from ten human cell types to perform a comparative analysis of super and stretch enhancers with respect to their chromatin profiles, cell type-specificity, and ability to control gene expression. We found that stretch enhancers are more abundant, more distal to transcription start sites, cover twice as much the genome, and are significantly less conserved than super-enhancers. In contrast, super-enhancers are significantly more enriched for active chromatin marks and cohesin complex, and more transcriptionally active than stretch enhancers. Importantly, a vast majority of super-enhancers (85%) overlap with only a small subset of stretch enhancers (13%), which are enriched for cell type-specific biological functions, and control cell identity genes. These results suggest that super-enhancers are transcriptionally more active and cell type-specific than stretch enhancers, and importantly, most of the stretch enhancers that are distinct from super-enhancers do not show an association with cell identity genes, are less active, and more likely to be poised enhancers.
Keywords: Gene regulation; epigenomics; genomics; stretch enhancers; super-enhancers.
Figures

References
-
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
