DNA methylation as a transcriptional regulator of the immune system
- PMID: 30170004
- PMCID: PMC6331288
- DOI: 10.1016/j.trsl.2018.08.001
DNA methylation as a transcriptional regulator of the immune system
Abstract
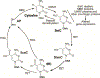
DNA methylation is a dynamic epigenetic modification with a prominent role in determining mammalian cell development, lineage identity, and transcriptional regulation. Primarily linked to gene silencing, novel technologies have expanded the ability to measure DNA methylation on a genome-wide scale and uncover context-dependent regulatory roles. The immune system is a prototypic model for studying how DNA methylation patterning modulates cell type- and stimulus-specific transcriptional programs. Preservation of host defense and organ homeostasis depends on fine-tuned epigenetic mechanisms controlling myeloid and lymphoid cell differentiation and function, which shape innate and adaptive immune responses. Dysregulation of these processes can lead to human immune system pathology as seen in blood malignancies, infections, and autoimmune diseases. Identification of distinct epigenotypes linked to pathogenesis carries the potential to validate therapeutic targets in disease prevention and management.
Copyright © 2018. Published by Elsevier Inc.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

