Heckman imputation models for binary or continuous MNAR outcomes and MAR predictors
- PMID: 30170561
- PMCID: PMC6119269
- DOI: 10.1186/s12874-018-0547-1
Heckman imputation models for binary or continuous MNAR outcomes and MAR predictors
Abstract
Background: Multiple imputation by chained equations (MICE) requires specifying a suitable conditional imputation model for each incomplete variable and then iteratively imputes the missing values. In the presence of missing not at random (MNAR) outcomes, valid statistical inference often requires joint models for missing observations and their indicators of missingness. In this study, we derived an imputation model for missing binary data with MNAR mechanism from Heckman's model using a one-step maximum likelihood estimator. We applied this approach to improve a previously developed approach for MNAR continuous outcomes using Heckman's model and a two-step estimator. These models allow us to use a MICE process and can thus also handle missing at random (MAR) predictors in the same MICE process.
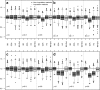
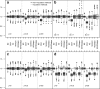
Methods: We simulated 1000 datasets of 500 cases. We generated the following missing data mechanisms on 30% of the outcomes: MAR mechanism, weak MNAR mechanism, and strong MNAR mechanism. We then resimulated the first three cases and added an additional 30% of MAR data on a predictor, resulting in 50% of complete cases. We evaluated and compared the performance of the developed approach to that of a complete case approach and classical Heckman's model estimates.
Results: With MNAR outcomes, only methods using Heckman's model were unbiased, and with a MAR predictor, the developed imputation approach outperformed all the other approaches.
Conclusions: In the presence of MAR predictors, we proposed a simple approach to address MNAR binary or continuous outcomes under a Heckman assumption in a MICE procedure.
Trial registration: ClinicalTrials.gov NCT00799760.
Keywords: Heckman’s model; Missing data; Missing not at random (MNAR); Multiple imputation by chained equation (MICE); Sample selection method.
Conflict of interest statement
Ethics approval and consent to participate
All the data have already been published in: “Efficacy of oseltamivir-zanamivir combination compared to each monotherapy for seasonal influenza: a randomized placebo-controlled trial.” (
Consent for publication
All the data have already been published in: “Efficacy of oseltamivir-zanamivir combination compared to each monotherapy for seasonal influenza: a randomized placebo-controlled trial.” (
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Little RJ, Rubin DB. Statistical Analysis with Missing Data. New York: Wiley; 2002.
-
- van Buuren S. Flexible Imputation of Missing Data. Boca Raton: CRC press; 2012.
-
- Fitzmaurice GM, Kenward MG, Molenberghs G, Verbeke G, Tsiatis AA. Handbook of Missing Data Methodology. Boca Raton: Chapman and Hall/CRC Press; 2014. Missing data: Introduction and statistical preliminaries.
-
- Little RJ. Pattern-mixture models for multivariate incomplete data. J Am Stat Assoc. 1993;88(421):125–34.
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

