Perturbed maintenance of transcriptional repression on the inactive X-chromosome in the mouse brain after Xist deletion
- PMID: 30170615
- PMCID: PMC6118007
- DOI: 10.1186/s13072-018-0219-8
Perturbed maintenance of transcriptional repression on the inactive X-chromosome in the mouse brain after Xist deletion
Abstract
Background: The long noncoding RNA Xist is critical for initiation and establishment of X-chromosome inactivation during embryogenesis in mammals, but it is unclear whether its continued expression is required for maintaining X-inactivation in vivo.
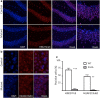
Results: By using an inactive X-chromosome-linked MeCP2-GFP reporter, which allowed us to enumerate reactivation events in the mouse brain even when they occur in very few cells, we found that deletion of Xist in the brain after establishment of X-chromosome inactivation leads to reactivation in 2-5% of neurons and in a smaller fraction of astrocytes. In contrast to global loss of both H3 lysine 27 trimethylation (H3K27m3) and histone H2A lysine 119 monoubiquitylation (H2AK119ub1) we observed upon Xist deletion, alterations in CpG methylation were subtle, and this was mirrored by only minor alterations in X-chromosome-wide gene expression levels, with highly expressed genes more prone to both derepression and demethylation compared to genes with low expression level.
Conclusion: Our results demonstrate that Xist plays a role in the maintenance of histone repressive marks, DNA methylation and transcriptional repression on the inactive X-chromosome, but that partial loss of X-dosage compensation in the absence of Xist in the brain is well tolerated.
Keywords: MeCP2; Noncoding RNA; Rett syndrome; X-chromosome inactivation; Xist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

