A Vegetable Fermentation Facility Hosts Distinct Microbiomes Reflecting the Production Environment
- PMID: 30171008
- PMCID: PMC6210109
- DOI: 10.1128/AEM.01680-18
A Vegetable Fermentation Facility Hosts Distinct Microbiomes Reflecting the Production Environment
Abstract
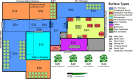
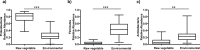
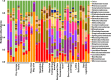
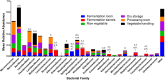
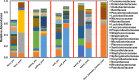
Fermented vegetables are highly popular internationally in part due to their enhanced nutritional properties, cultural history, and desirable sensorial properties. In some instances, fermented foods provide a rich source of the beneficial microbial communities that could promote gastrointestinal health. The indigenous microbiota that colonize fermentation facilities may impact food quality, food safety, and spoilage risks and maintain the nutritive value of the product. Here, microbiomes within sauerkraut production facilities were profiled to characterize variance across surfaces and to determine the sources of these bacteria. Accordingly, we used high-throughput sequencing of the 16S rRNA gene in combination with whole-genome shotgun analyses to explore biogeographical patterns of microbial diversity and assembly within the production facility. Our results indicate that raw cabbage and vegetable handling surfaces exhibit more similar microbiomes relative to the fermentation room, processing area, and dry storage surfaces. We identified biomarker bacterial phyla and families that are likely to originate from the raw cabbage and vegetable handling surfaces. Raw cabbage was identified as the main source of bacteria to seed the facility, with human handling contributing a minor source of inoculation. Leuconostoc and Lactobacillaceae dominated all surfaces where spontaneous fermentation occurs, as these taxa are associated with the process. Wall, floor, ceiling, and barrel surfaces host unique microbial signatures. This study demonstrates that diverse bacterial communities are widely distributed within the production facility and that these communities assemble nonrandomly, depending on the surface type.IMPORTANCE Fermented vegetables play a major role in global food systems and are widely consumed by various global cultures. In this study, we investigated an industrial facility that produces spontaneous fermented sauerkraut without the aid of starter cultures. This provides a unique system to explore and track the origins of an "in-house" microbiome in an industrial environment. Raw vegetables and the surfaces on which they are handled were identified as the likely source of bacterial communities rather than human contamination. As fermented vegetables increase in popularity on a global scale, understanding their production environment may help maintain quality and safety goals.
Keywords: fermentation; food microbiology; lactic acid bacteria; microbiology of the built environment; microbiome; phylogenetic diversity.
Copyright © 2018 American Society for Microbiology.
Figures




References
-
- Pederson CS, Albury MN. 1969. Bulletin: number 824: the sauerkraut fermentation, Agricultural Experiment Station, Geneva, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

