Precision and accuracy of single-molecule FRET measurements-a multi-laboratory benchmark study
- PMID: 30171252
- PMCID: PMC6121742
- DOI: 10.1038/s41592-018-0085-0
Precision and accuracy of single-molecule FRET measurements-a multi-laboratory benchmark study
Erratum in
-
Publisher Correction: Precision and accuracy of single-molecule FRET measurements-a multi-laboratory benchmark study.Nat Methods. 2018 Nov;15(11):984. doi: 10.1038/s41592-018-0193-x. Nat Methods. 2018. PMID: 30327572 Free PMC article.
Abstract
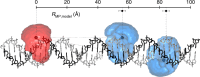
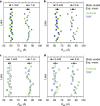
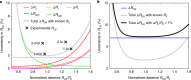
Single-molecule Förster resonance energy transfer (smFRET) is increasingly being used to determine distances, structures, and dynamics of biomolecules in vitro and in vivo. However, generalized protocols and FRET standards to ensure the reproducibility and accuracy of measurements of FRET efficiencies are currently lacking. Here we report the results of a comparative blind study in which 20 labs determined the FRET efficiencies (E) of several dye-labeled DNA duplexes. Using a unified, straightforward method, we obtained FRET efficiencies with s.d. between ±0.02 and ±0.05. We suggest experimental and computational procedures for converting FRET efficiencies into accurate distances, and discuss potential uncertainties in the experiment and the modeling. Our quantitative assessment of the reproducibility of intensity-based smFRET measurements and a unified correction procedure represents an important step toward the validation of distance networks, with the ultimate aim of achieving reliable structural models of biomolecular systems by smFRET-based hybrid methods.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Förster T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann. Phys. 1948;437:55–75.
-
- Murchie AI, et al. Fluorescence energy transfer shows that the four-way DNA junction is a right-handed cross of antiparallel molecules. Nature. 1989;341:763–766. - PubMed
-
- Mekler V, et al. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. - PubMed

