Assembling the Community-Scale Discoverable Human Proteome
- PMID: 30172843
- PMCID: PMC6279426
- DOI: 10.1016/j.cels.2018.08.004
Assembling the Community-Scale Discoverable Human Proteome
Abstract
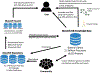
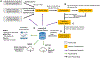
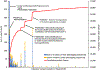
The increasing throughput and sharing of proteomics mass spectrometry data have now yielded over one-third of a million public mass spectrometry runs. However, these discoveries are not continuously aggregated in an open and error-controlled manner, which limits their utility. To facilitate the reusability of these data, we built the MassIVE Knowledge Base (MassIVE-KB), a community-wide, continuously updating knowledge base that aggregates proteomics mass spectrometry discoveries into an open reusable format with full provenance information for community scrutiny. Reusing >31 TB of public human data stored in a mass spectrometry interactive virtual environment (MassIVE), the MassIVE-KB contains >2.1 million precursors from 19,610 proteins (48% larger than before; 97% of the total) and doubles proteome coverage to 6 million amino acids (54% of the proteome) with strict library-scale false discovery controls, thereby providing evidence for 430 proteins for which sufficient protein-level evidence was previously missing. Furthermore, MassIVE-KB can inform experimental design, helps identify and quantify new data, and provides tools for community construction of specialized spectral libraries.
Keywords: algorithms; big data; knowledge base; proteomics; repositories; spectral libraries; tandem mass spectrometry.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
N.B. was a co-founder, had an equity interest, and received income from Digital Proteomics, LLC through 2017. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies. Digital Proteomics was not involved in the research presented here.
Figures


Comment in
-
Proteomics data reuse with MassIVE-KB.Nat Methods. 2019 Jan;16(1):26. doi: 10.1038/s41592-018-0283-9. Nat Methods. 2019. PMID: 30573830 No abstract available.
References
-
- Boutet E, Lieberherr D, Tognolli M, Schneider M, Bansal P, Bridge AJ, Poux S, Bougueleret L, and Xenarios I (2016). Uniprotkb/swiss-prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol. Biol 1374, 23–54. - PubMed
-
- Craig R, Cortens JC, Fenyo D, and Beavis RC (2006). Using annotated peptide mass spectrum libraries for protein identification. J. Proteome Res 5, 1843–1849. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

