PubCaseFinder: A Case-Report-Based, Phenotype-Driven Differential-Diagnosis System for Rare Diseases
- PMID: 30173820
- PMCID: PMC6128307
- DOI: 10.1016/j.ajhg.2018.08.003
PubCaseFinder: A Case-Report-Based, Phenotype-Driven Differential-Diagnosis System for Rare Diseases
Abstract
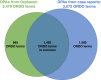
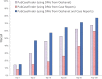
Recently, to speed up the differential-diagnosis process based on symptoms and signs observed from an affected individual in the diagnosis of rare diseases, researchers have developed and implemented phenotype-driven differential-diagnosis systems. The performance of those systems relies on the quantity and quality of underlying databases of disease-phenotype associations (DPAs). Although such databases are often developed by manual curation, they inherently suffer from limited coverage. To address this problem, we propose a text-mining approach to increase the coverage of DPA databases and consequently improve the performance of differential-diagnosis systems. Our analysis showed that a text-mining approach using one million case reports obtained from PubMed could increase the coverage of manually curated DPAs in Orphanet by 125.6%. We also present PubCaseFinder (see Web Resources), a new phenotype-driven differential-diagnosis system in a freely available web application. By utilizing automatically extracted DPAs from case reports in addition to manually curated DPAs, PubCaseFinder improves the performance of automated differential diagnosis. Moreover, PubCaseFinder helps clinicians search for relevant case reports by using phenotype-based comparisons and confirm the results with detailed contextual information.
Keywords: Human Phenotype Ontology; PubCaseFinder; case report; differential diagnosis; disease-phenotype association; rare disease.
Copyright © 2018 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures







Similar articles
-
Advances in the development of PubCaseFinder, including the new application programming interface and matching algorithm.Hum Mutat. 2022 Jun;43(6):734-742. doi: 10.1002/humu.24341. Epub 2022 Feb 22. Hum Mutat. 2022. PMID: 35143083 Free PMC article.
-
Rare disease knowledge enrichment through a data-driven approach.BMC Med Inform Decis Mak. 2019 Feb 14;19(1):32. doi: 10.1186/s12911-019-0752-9. BMC Med Inform Decis Mak. 2019. PMID: 30764825 Free PMC article.
-
PhenoDis: a comprehensive database for phenotypic characterization of rare cardiac diseases.Orphanet J Rare Dis. 2018 Jan 25;13(1):22. doi: 10.1186/s13023-018-0765-y. Orphanet J Rare Dis. 2018. PMID: 29370821 Free PMC article.
-
[Rare-disease data standards].Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2022 Nov;65(11):1126-1132. doi: 10.1007/s00103-022-03591-2. Epub 2022 Sep 23. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2022. PMID: 36149471 Free PMC article. Review. German.
-
Meeting Patients' Right to the Correct Diagnosis: Ongoing International Initiatives on Undiagnosed Rare Diseases and Ethical and Social Issues.Int J Environ Res Public Health. 2018 Sep 21;15(10):2072. doi: 10.3390/ijerph15102072. Int J Environ Res Public Health. 2018. PMID: 30248891 Free PMC article. Review.
Cited by
-
Generalizability of Polygenic Risk Scores for Breast Cancer Among Women With European, African, and Latinx Ancestry.JAMA Netw Open. 2021 Aug 2;4(8):e2119084. doi: 10.1001/jamanetworkopen.2021.19084. JAMA Netw Open. 2021. PMID: 34347061 Free PMC article.
-
PhenoApt leverages clinical expertise to prioritize candidate genes via machine learning.Am J Hum Genet. 2022 Feb 3;109(2):270-281. doi: 10.1016/j.ajhg.2021.12.008. Epub 2022 Jan 20. Am J Hum Genet. 2022. PMID: 35063063 Free PMC article.
-
Visualizing the phenotype diversity: a case study of Alexander disease.Genomics Inform. 2021 Sep;19(3):e28. doi: 10.5808/gi.21016. Epub 2021 Sep 30. Genomics Inform. 2021. PMID: 34638175 Free PMC article.
-
Diagnosis support systems for rare diseases: a scoping review.Orphanet J Rare Dis. 2020 Apr 16;15(1):94. doi: 10.1186/s13023-020-01374-z. Orphanet J Rare Dis. 2020. PMID: 32299466 Free PMC article.
-
Rare, rarer, it still has not happened.Croat Med J. 2019 Dec 31;60(6):565-569. doi: 10.3325/cmj.2019.60.565. Croat Med J. 2019. PMID: 31894924 Free PMC article. No abstract available.
References
-
- Boat T.F., Field M.J., editors. Rare diseases and orphan products: Accelerating research and development. National Academies Press; 2011. - PubMed
-
- Sawyer S.L., Hartley T., Dyment D.A., Beaulieu C.L., Schwartzentruber J., Smith A., Bedford H.M., Bernard G., Bernier F.P., Brais B., FORGE Canada Consortium. Care4Rare Canada Consortium Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: Time to address gaps in care. Clin. Genet. 2016;89:275–284. - PMC - PubMed
-
- Stranneheim H., Wedell A. Exome and genome sequencing: A revolution for the discovery and diagnosis of monogenic disorders. J. Intern. Med. 2016;279:3–15. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

