Capsid-CPSF6 Interaction Licenses Nuclear HIV-1 Trafficking to Sites of Viral DNA Integration
- PMID: 30173955
- PMCID: PMC6368089
- DOI: 10.1016/j.chom.2018.08.002
Capsid-CPSF6 Interaction Licenses Nuclear HIV-1 Trafficking to Sites of Viral DNA Integration
Abstract
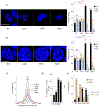
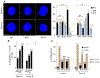
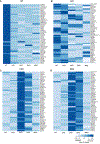
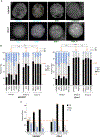
HIV-1 integration into the host genome favors actively transcribed genes. Prior work indicated that the nuclear periphery provides the architectural basis for integration site selection, with viral capsid-binding host cofactor CPSF6 and viral integrase-binding cofactor LEDGF/p75 contributing to selection of individual sites. Here, by investigating the early phase of infection, we determine that HIV-1 traffics throughout the nucleus for integration. CPSF6-capsid interactions allow the virus to bypass peripheral heterochromatin and penetrate the nuclear structure for integration. Loss of interaction with CPSF6 dramatically alters virus localization toward the nuclear periphery and integration into transcriptionally repressed lamina-associated heterochromatin, while loss of LEDGF/p75 does not significantly affect intranuclear HIV-1 localization. Thus, CPSF6 serves as a master regulator of HIV-1 intranuclear localization by trafficking viral preintegration complexes away from heterochromatin at the periphery toward gene-dense chromosomal regions within the nuclear interior.
Keywords: CPSF6; HIV capsid; HIV integrase; HIV integration; HIV trafficking; LEDGF/p75; lamina-associated domain; nuclear trafficking.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures


References
-
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, and Bickmore WA (2001). The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet 10, 211–219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

