Organoids from Nephrotic Disease-Derived iPSCs Identify Impaired NEPHRIN Localization and Slit Diaphragm Formation in Kidney Podocytes
- PMID: 30174315
- PMCID: PMC6135868
- DOI: 10.1016/j.stemcr.2018.08.003
Organoids from Nephrotic Disease-Derived iPSCs Identify Impaired NEPHRIN Localization and Slit Diaphragm Formation in Kidney Podocytes
Abstract
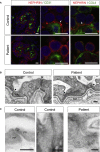
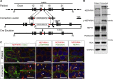
Mutations in the NPHS1 gene, which encodes NEPHRIN, cause congenital nephrotic syndrome, resulting from impaired slit diaphragm (SD) formation in glomerular podocytes. However, methods for SD reconstitution have been unavailable, thereby limiting studies in the field. In the present study, we established human induced pluripotent stem cells (iPSCs) from a patient with an NPHS1 missense mutation, and reproduced the SD formation process using iPSC-derived kidney organoids. The mutant NEPHRIN failed to become localized on the cell surface for pre-SD domain formation in the induced podocytes. Upon transplantation, the mutant podocytes developed foot processes, but exhibited impaired SD formation. Genetic correction of the single amino acid mutation restored NEPHRIN localization and phosphorylation, colocalization of other SD-associated proteins, and SD formation. Thus, these kidney organoids from patient-derived iPSCs identified SD abnormalities in the podocytes at the initial phase of congenital nephrotic disease.
Keywords: NEPHRIN; NPHS1; iPSCs; kidney; nephrotic syndrome; podocyte; slit diaphragm.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Beltcheva O., Martin P., Lenkkeri U., Tryggvason K. Mutation spectrum in the nephrin gene (NPHS1) in congenital nephrotic syndrome. Hum. Mutat. 2001;17:368–373. - PubMed
-
- Colls P., Blanco J., Martínez-Pasarell O., Vidal F., Egozcue J., Márquez C., Guitart M., Templado C. Chromosome segregation in a man heterozygous for a pericentric inversion, inv(9)(p11q13), analyzed by using sperm karyotyping and two-color fluorescence in situ hybridization on sperm nuclei. Hum. Genet. 1997;99:761–765. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

