A two-stage hidden Markov model design for biomarker detection, with application to microbiome research
- PMID: 30174757
- PMCID: PMC6116560
- DOI: 10.1007/s12561-017-9187-y
A two-stage hidden Markov model design for biomarker detection, with application to microbiome research
Abstract
It has been recognized that for appropriately ordered data, hidden Markov models (HMM) with local false discovery rate (FDR) control can increase the power to detect significant associations. For many high-throughput technologies, the cost still limits their application. Two-stage designs are attractive, in which a set of interesting features or biomarkers is identified in a first stage, and then followed up in a second stage. However, to our knowledge no two-stage FDR control with HMMs has been developed. In this paper, we study an efficient HMM-FDR based two-stage design, using a simple integrated analysis procedure across the stages. Numeric studies show its excellent performance when compared to available methods. A power analysis method is also proposed. We use examples from microbiome data to illustrate the methods.
Keywords: Biomarker; False discovery rates; Hidden Markov model; Metagenomics; Metatranscriptomics; PCR.
Figures



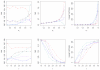

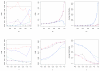
References
-
- Zehetmayer S, Bauer P, Posch M. Two-stage designs for experiments with a large number of hypotheses. Bioinformatics. 2005;21:3771–3777. - PubMed
-
- Breslow NE, Cain KC. Logistic regression for two-stage case-control data. Biometrika. 1988;71:11–20.
-
- Goll A, Bauer P. Two-stage designs applying methods differing in costs. Bioinformatics. 2007;23:1519–26. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
