Limitations of Correlation-Based Inference in Complex Virus-Microbe Communities
- PMID: 30175237
- PMCID: PMC6113591
- DOI: 10.1128/mSystems.00084-18
Limitations of Correlation-Based Inference in Complex Virus-Microbe Communities
Abstract
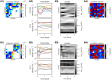
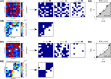
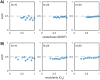
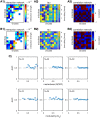
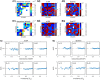
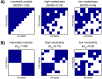
Microbes are present in high abundances in the environment and in human-associated microbiomes, often exceeding 1 million per ml. Viruses of microbes are present in even higher abundances and are important in shaping microbial populations, communities, and ecosystems. Given the relative specificity of viral infection, it is essential to identify the functional linkages between viruses and their microbial hosts, particularly given dynamic changes in virus and host abundances. Multiple approaches have been proposed to infer infection networks from time series of in situ communities, among which correlation-based approaches have emerged as the de facto standard. In this work, we evaluate the accuracy of correlation-based inference methods using an in silico approach. In doing so, we compare predicted networks to actual networks to assess the self-consistency of correlation-based inference. At odds with assumptions underlying its widespread use, we find that correlation is a poor predictor of interactions in the context of viral infection and lysis of microbial hosts. The failure to predict interactions holds for methods that leverage product-moment, time-lagged, and relative-abundance-based correlations. In closing, we discuss alternative inference methods, particularly model-based methods, as a means to infer interactions in complex microbial communities with viruses. IMPORTANCE Inferring interactions from population time series is an active and ongoing area of research. It is relevant across many biological systems-particularly in virus-microbe communities, but also in gene regulatory networks, neural networks, and ecological communities broadly. Correlation-based inference-using correlations to predict interactions-is widespread. However, it is well-known that "correlation does not imply causation." Despite this, many studies apply correlation-based inference methods to experimental time series without first assessing the potential scope for accurate inference. Here, we find that several correlation-based inference methods fail to recover interactions within in silico virus-microbe communities, raising questions on their relevance when applied in situ.
Keywords: correlation; inference; interaction network; microbial ecology; viral ecology.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
