Plastome characteristics of Cannabaceae
- PMID: 30175293
- PMCID: PMC6114266
- DOI: 10.1016/j.pld.2018.04.003
Plastome characteristics of Cannabaceae
Abstract
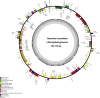
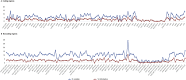
Cannabaceae is an economically important family that includes ten genera and ca. 117 accepted species. To explore the structure and size variation of their plastomes, we sequenced ten plastomes representing all ten genera of Cannabaceae. Each plastome possessed the typical angiosperm quadripartite structure and contained a total of 128 genes. The Inverted Repeat (IR) regions in five plastomes had experienced small expansions (330-983 bp) into the Large Single-Copy (LSC) region. The plastome of Chaetachme aristata has experienced a 942-bp IR contraction and lost rpl22 and rps19 in its IRs. The substitution rates of rps19 and rpl22 decreased after they shifted from the LSC to IR. A 270-bp inversion was detected in the Parasponia rugosa plastome, which might have been mediated by 18-bp inverted repeats. Repeat sequences, simple sequence repeats, and nucleotide substitution rates varied among these plastomes. Molecular markers with more than 13% variable sites and 5% parsimony-informative sites were identified, which may be useful for further phylogenetic analysis and species identification. Our results show strong support for a sister relationship between Gironniera and Lozanell (BS = 100). Celtis, Cannabis-Humulus, Chaetachme-Pteroceltis, and Trema-Parasponia formed a strongly supported clade, and their relationships were well resolved with strong support (BS = 100). The availability of these ten plastomes provides valuable genetic information for accurately identifying species, clarifying taxonomy and reconstructing the intergeneric phylogeny of Cannabaceae.
Keywords: IR expansion/contraction; Phylogenomics; Plastome; Repeats; SSR; Sequence divergence.
Figures





Similar articles
-
Plastid phylogenomic insights into the evolution of subfamily Dialioideae (Leguminosae).Plant Divers. 2020 Jul 15;43(1):27-34. doi: 10.1016/j.pld.2020.06.008. eCollection 2021 Feb. Plant Divers. 2020. PMID: 33778222 Free PMC article.
-
Plastome Evolution and Phylogeny of Orchidaceae, With 24 New Sequences.Front Plant Sci. 2020 Feb 21;11:22. doi: 10.3389/fpls.2020.00022. eCollection 2020. Front Plant Sci. 2020. PMID: 32153600 Free PMC article.
-
Total duplication of the small single copy region in the angiosperm plastome: Rearrangement and inverted repeat instability in Asarum.Am J Bot. 2018 Jan;105(1):71-84. doi: 10.1002/ajb2.1001. Epub 2018 Feb 12. Am J Bot. 2018. PMID: 29532923
-
Complete plastome sequence of Iodes cirrhosa Turcz., the first in the Icacinaceae, comparative genomic analyses and possible split of Idoes species in response to climate changes.PeerJ. 2019 Apr 1;7:e6663. doi: 10.7717/peerj.6663. eCollection 2019. PeerJ. 2019. PMID: 30972252 Free PMC article.
-
A comparative analysis of plastome evolution in autotrophic Piperales.Am J Bot. 2024 Mar;111(3):e16300. doi: 10.1002/ajb2.16300. Epub 2024 Mar 12. Am J Bot. 2024. PMID: 38469876 Review.
Cited by
-
Development of genomic resources for the genus Celtis (Cannabaceae) based on genome skimming data.Plant Divers. 2020 Oct 5;43(1):43-53. doi: 10.1016/j.pld.2020.09.005. eCollection 2021 Feb. Plant Divers. 2020. PMID: 33778224 Free PMC article.
-
SilicoDArT and SNP markers for genetic diversity and population structure analysis of Trema orientalis; a fodder species.PLoS One. 2022 Aug 22;17(8):e0267464. doi: 10.1371/journal.pone.0267464. eCollection 2022. PLoS One. 2022. PMID: 35994436 Free PMC article.
-
The complete chloroplast genome sequence of Berchemia racemosa Siebold & Zucc. (Rhamnaceae), a rare plant species in Korea.Mitochondrial DNA B Resour. 2023 Jan 2;8(1):69-72. doi: 10.1080/23802359.2022.2161329. eCollection 2023. Mitochondrial DNA B Resour. 2023. PMID: 36620322 Free PMC article.
-
New Insights Into the Plastome Evolution of the Millettioid/Phaseoloid Clade (Papilionoideae, Leguminosae).Front Plant Sci. 2020 Mar 10;11:151. doi: 10.3389/fpls.2020.00151. eCollection 2020. Front Plant Sci. 2020. PMID: 32210983 Free PMC article.
-
Cannabinoids from inflorescences fractions of Trema orientalis (L.) Blume (Cannabaceae) against human pathogenic bacteria.PeerJ. 2021 May 13;9:e11446. doi: 10.7717/peerj.11446. eCollection 2021. PeerJ. 2021. PMID: 34035994 Free PMC article.
References
-
- Altschul S.F., Gish W., Miller W. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Bell C.D., Soltis D.E., Soltis P.S. The age and diversification of the angiosperms re-revisited. Am. J. Bot. 2010;97:1296–1303. - PubMed
-
- Byng J.W., Chase M.W., Christenhusz M.J.M. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016;181:1–20.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

