Covalent Immobilization of Proteins for the Single Molecule Force Spectroscopy
- PMID: 30176022
- PMCID: PMC6128213
- DOI: 10.3791/58167
Covalent Immobilization of Proteins for the Single Molecule Force Spectroscopy
Abstract
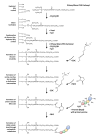
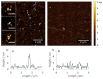
In recent years, atomic force microscopy (AFM) based single molecule force spectroscopy (SMFS) extended our understanding of molecular properties and functions. It gave us the opportunity to explore a multiplicity of biophysical mechanisms, e.g., how bacterial adhesins bind to host surface receptors in more detail. Among other factors, the success of SMFS experiments depends on the functional and native immobilization of the biomolecules of interest on solid surfaces and AFM tips. Here, we describe a straightforward protocol for the covalent coupling of proteins to silicon surfaces using silane-PEG-carboxyls and the well-established N-hydroxysuccinimid/1-ethyl-3-(3-dimethyl-aminopropyl)carbodiimid (EDC/NHS) chemistry in order to explore the interaction of pilus-1 adhesin RrgA from the Gram-positive bacterium Streptococcus pneumoniae (S. pneumoniae) with the extracellular matrix protein fibronectin (Fn). Our results show that the surface functionalization leads to a homogenous distribution of Fn on the glass surface and to an appropriate concentration of RrgA on the AFM cantilever tip, apparent by the target value of up to 20% of interaction events during SMFS measurements and revealed that RrgA binds to Fn with a mean force of 52 pN. The protocol can be adjusted to couple via site specific free thiol groups. This results in a predefined protein or molecule orientation and is suitable for other biophysical applications besides the SMFS.
References
-
- Binnig G, Quate CF, Gerber C. Atomic Force Microscope. Physical Review Letters. 1986;56:930–933. - PubMed
-
- Muller DJ, Dufrene YF. Atomic Force Microscopy as a Multifunctional Molecular Toolbox in Nanobiotechnology. Nature Nanotechnology. 2008;3(5):261–269. - PubMed
-
- Hinterdorfer P, Dufrene YF. Detection and Localization of Single Molecular Recognition Events Using Atomic Force Microscopy. Nature Methods. 2006;3(5):347–355. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous