Parallel damage in mitochondria and lysosomes is an efficient way to photoinduce cell death
- PMID: 30176156
- PMCID: PMC6333451
- DOI: 10.1080/15548627.2018.1515609
Parallel damage in mitochondria and lysosomes is an efficient way to photoinduce cell death
Abstract
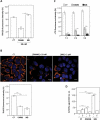
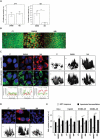
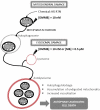
Cells challenged by photosensitized oxidations face strong redox stresses and rely on autophagy to either survive or die. However, the use of macroautophagy/autophagy to improve the efficiency of photosensitizers, in terms of inducing cell death, remains unexplored. Here, we addressed the concept that a parallel damage in the membranes of mitochondria and lysosomes leads to a scenario of autophagy malfunction that can greatly improve the efficiency of the photosensitizer to cause cell death. Specific damage to these organelles was induced by irradiation of cells pretreated with 2 phenothiazinium salts, methylene blue (MB) and 1,9-dimethyl methylene blue (DMMB). At a low concentration level (10 nM), only DMMB could induce mitochondrial damage, leading to mitophagy activation, which did not progress to completion because of the parallel damage in lysosome, triggering cell death. MB-induced photodamage was perceived almost instantaneously after irradiation, in response to a massive and nonspecific oxidative stress at a higher concentration range (2 µM). We showed that the parallel damage in mitochondria and lysosomes activates and inhibits mitophagy, leading to a late and more efficient cell death, offering significant advantage (2 orders of magnitude) over photosensitizers that cause unspecific oxidative stress. We are confident that this concept can be used to develop better light-activated drugs. Abbreviations: ΔΨm: mitochondrial transmembrane inner potential; AAU: autophagy arbitrary units; ATG5, autophagy related 5; ATG7: autophagy related 7; BAF: bafilomycin A1; BSA: bovine serum albumin; CASP3: caspase 3; CF: carboxyfluorescein; CTSB: cathepsin B; CVS: crystal violet staining; DCF: dichlorofluorescein; DCFH2: 2',7'-dichlorodihydrofluorescein; DMMB: 1,9-dimethyl methylene blue; ER: endoplasmic reticulum; HaCaT: non-malignant immortal keratinocyte cell line from adult human skin; HP: hydrogen peroxide; LC3B-II: microtubule associated protein 1 light chain 3 beta-II; LMP: lysosomal membrane permeabilization; LTG: LysoTracker™ Green DND-26; LTR: LysoTracker™ Red DND-99; 3-MA: 3-methyladenine; MB: methylene blue; mtDNA: mitochondrial DNA; MitoSOX™: red mitochondrial superoxide probe; MTDR: MitoTracker™ Deep Red FM; MTO: MitoTracker™ Orange CMTMRos; MT-ND1: mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 1; MTT: methylthiazolyldiphenyl-tetrazolium bromide; 1O2: singlet oxygen; OH. hydroxil radical; PRKN/parkin: parkin RBR E3 ubiquitin protein ligase; PBS: phosphate-buffered saline; PI: propidium iodide; PDT: photodynamic therapy; PS: photosensitizer; QPCR: gene-specific quantitative PCR-based; Rh123: rhodamine 123; ROS: reactive oxygen species RTN: rotenone; SQSTM1/p62: sequestosome 1; SUVs: small unilamellar vesicles; TBS: Tris-buffered saline.
Keywords: Cell death; membrane damage; methylene blue; mitophagy; phenothiazines; photodynamic efficiency.
Figures





Similar articles
-
HIV-1 TAT-mediated microglial activation: role of mitochondrial dysfunction and defective mitophagy.Autophagy. 2018;14(9):1596-1619. doi: 10.1080/15548627.2018.1476810. Epub 2018 Jul 26. Autophagy. 2018. PMID: 29966509 Free PMC article.
-
AT 101 induces early mitochondrial dysfunction and HMOX1 (heme oxygenase 1) to trigger mitophagic cell death in glioma cells.Autophagy. 2018;14(10):1693-1709. doi: 10.1080/15548627.2018.1476812. Epub 2018 Jul 21. Autophagy. 2018. PMID: 29938581 Free PMC article.
-
Mitochondrial respiratory chain deficiency inhibits lysosomal hydrolysis.Autophagy. 2019 Sep;15(9):1572-1591. doi: 10.1080/15548627.2019.1586256. Epub 2019 Mar 27. Autophagy. 2019. PMID: 30917721 Free PMC article.
-
Organelle-specific autophagy in inflammatory diseases: a potential therapeutic target underlying the quality control of multiple organelles.Autophagy. 2021 Feb;17(2):385-401. doi: 10.1080/15548627.2020.1725377. Epub 2020 Feb 12. Autophagy. 2021. PMID: 32048886 Free PMC article. Review.
-
Assessing autophagy in the context of photodynamic therapy.Autophagy. 2010 Jan;6(1):7-18. doi: 10.4161/auto.6.1.10220. Epub 2010 Jan 1. Autophagy. 2010. PMID: 19855190 Free PMC article. Review.
Cited by
-
Serum Nitric Oxide Level Serves as a Potential Prognostic Biomarker in ACLF Patients.Int J Gen Med. 2022 Aug 22;15:6713-6723. doi: 10.2147/IJGM.S379837. eCollection 2022. Int J Gen Med. 2022. PMID: 36034183 Free PMC article.
-
Cyanine dyes in the mitochondria-targeting photodynamic and photothermal therapy.Commun Chem. 2024 Aug 13;7(1):180. doi: 10.1038/s42004-024-01256-6. Commun Chem. 2024. PMID: 39138299 Free PMC article. Review.
-
Static Magnetic Field Reduces Intracellular ROS Levels and Protects Cells Against Peroxide-Induced Damage: Suggested Roles for Catalase.Neurotox Res. 2023 Dec 14;42(1):2. doi: 10.1007/s12640-023-00679-8. Neurotox Res. 2023. PMID: 38095761
-
Progress in the photodynamic therapy treatment of Leishmaniasis.Braz J Med Biol Res. 2021 Oct 29;54(12):e11570. doi: 10.1590/1414-431X2021e11570. eCollection 2021. Braz J Med Biol Res. 2021. PMID: 34730683 Free PMC article. Review.
-
Oxidative Stress Enhances Autophagy-Mediated Death Of Stem Cells Through Erk1/2 Signaling Pathway - Implications For Neurotransplantations.Stem Cell Rev Rep. 2021 Dec;17(6):2347-2358. doi: 10.1007/s12015-021-10212-z. Epub 2021 Sep 6. Stem Cell Rev Rep. 2021. PMID: 34487308
References
-
- Lemasters JJ, Autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
