RpfC regulates the expression of the key regulator hrpX of the hrp/T3SS system in Xanthomonas campestris pv. campestris
- PMID: 30176800
- PMCID: PMC6122198
- DOI: 10.1186/s12866-018-1233-5
RpfC regulates the expression of the key regulator hrpX of the hrp/T3SS system in Xanthomonas campestris pv. campestris
Abstract
Background: The Gram-negative phytopathogenic bacterium Xanthomonas campestris pv. campestris recruits the hrp/T3SS system to inject pathogenicity effector proteins into host cells and uses the rpf/DSF cell-cell signaling system to regulate the expression of virulence factors such as extracellular enzymes and polysaccharide. Whether these two systems have any connection is unknown.
Methods: Positive regulator candidates affecting hrpX expression were identified by sacB strategy. The transcriptional expression was determined by qRT-PCR and GUS activity analysis. Transcriptome analysis was performed by RNA deep-sequencing. The hypersensitive response (HR) was determined in the nonhost plant pepper ECW-10R and electrolyte leakage assay.
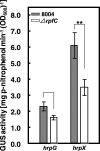
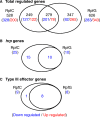
Results: Mutation of the gene encoding the sensor RpfC of the rpf/DSF system significantly reduced the expression of hrpX, the key regulator of the hrp/T3SS system, all of the genes in the hrp cluster and most reported type III effector genes. Mutation of rpfG did not affect the expression of hrpX. The rpfC mutant showed a delayed and weakened HR induction.
Conclusions: RpfC positively regulates the expression of hrpX independent of RpfG, showing a complex regulatory network linking the rpf/DSF and hrp/T3SS systems.
Keywords: RpfC; Xanthomonas; hrpX.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




Similar articles
-
[Identification of a new type III effector XC3176 in Xanthomonas campestris pv. campestris].Wei Sheng Wu Xue Bao. 2015 Oct 4;55(10):1264-72. Wei Sheng Wu Xue Bao. 2015. PMID: 26939454 Chinese.
-
The Zur of Xanthomonas campestris is involved in hypersensitive response and positively regulates the expression of the hrp cluster via hrpX but not hrpG.Mol Plant Microbe Interact. 2009 Mar;22(3):321-9. doi: 10.1094/MPMI-22-3-0321. Mol Plant Microbe Interact. 2009. PMID: 19245326
-
Identification of six type III effector genes with the PIP box in Xanthomonas campestris pv. campestris and five of them contribute individually to full pathogenicity.Mol Plant Microbe Interact. 2009 Nov;22(11):1401-11. doi: 10.1094/MPMI-22-11-1401. Mol Plant Microbe Interact. 2009. PMID: 19810809
-
Cell-cell signaling, cyclic di-GMP turnover and regulation of virulence in Xanthomonas campestris.Res Microbiol. 2006 Dec;157(10):899-904. doi: 10.1016/j.resmic.2006.08.001. Epub 2006 Sep 18. Res Microbiol. 2006. PMID: 17008065 Review.
-
Getting across--bacterial type III effector proteins on their way to the plant cell.EMBO J. 2002 Oct 15;21(20):5313-22. doi: 10.1093/emboj/cdf536. EMBO J. 2002. PMID: 12374732 Free PMC article. Review.
Cited by
-
Flp, a Fis-like protein, contributes to the regulation of type III secretion and virulence processes in the phytopathogen Xanthomonas campestris pv. campestris.Mol Plant Pathol. 2019 Aug;20(8):1119-1133. doi: 10.1111/mpp.12818. Epub 2019 May 14. Mol Plant Pathol. 2019. PMID: 31090173 Free PMC article.
-
Identification of c-di-GMP Signaling Components in Xanthomonas oryzae and Their Orthologs in Xanthomonads Involved in Regulation of Bacterial Virulence Expression.Front Microbiol. 2019 Jul 11;10:1402. doi: 10.3389/fmicb.2019.01402. eCollection 2019. Front Microbiol. 2019. PMID: 31354637 Free PMC article. Review.
-
The HrpG/HrpX Regulon of Xanthomonads-An Insight to the Complexity of Regulation of Virulence Traits in Phytopathogenic Bacteria.Microorganisms. 2021 Jan 16;9(1):187. doi: 10.3390/microorganisms9010187. Microorganisms. 2021. PMID: 33467109 Free PMC article. Review.
-
Two Non-Necrotic Disease Resistance Types Distinctly Affect the Expression of Key Pathogenic Determinants of Xanthomonas euvesicatoria in Pepper.Plants (Basel). 2022 Dec 24;12(1):89. doi: 10.3390/plants12010089. Plants (Basel). 2022. PMID: 36616218 Free PMC article.
-
Evolution of two-component quorum sensing systems.Access Microbiol. 2022 Jan 12;4(1):000303. doi: 10.1099/acmi.0.000303. eCollection 2022. Access Microbiol. 2022. PMID: 35252749 Free PMC article.
References
-
- Vorhölter FJ, Schneiker S, Goesmann A, Krause L, Bekel T, Kaiser O, et al. The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J Biotechnol. 2008;134(1–2):33–45. doi: 10.1016/j.jbiotec.2007.12.013. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

