Genotyping and phylogenetic placement of Bacillus anthracis isolates from Finland, a country with rare anthrax cases
- PMID: 30176810
- PMCID: PMC6122712
- DOI: 10.1186/s12866-018-1250-4
Genotyping and phylogenetic placement of Bacillus anthracis isolates from Finland, a country with rare anthrax cases
Abstract
Background: Anthrax, the zoonotic disease caused by the gram-positive bacterium Bacillus anthracis, is nowadays rare in northern parts of Europe including Finland and Scandinavia. Only two minor outbreaks of anthrax in 1988 and in 2004 and one sporadic infection in 2008 have been detected in animals in Finland since the 1970's. Here, we report on two Finnish B. anthracis strains that were isolated from spleen and liver of a diseased calf related to the outbreak in 1988 (strain HKI4363/88) and from a local scrotum and testicle infection of a bull in 2008 (strain BA2968). These infections occurred in two rural Finnish regions, i.e., Ostrobothnia in western Finland and Päijänne Tavastia in southern Finland, respectively.
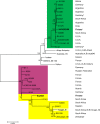
Results: The isolates were genetically characterized by PCR-based methods such as multilocus variable number of tandem repeat analysis (MLVA) and whole genome-sequence analysis (WGS). Phylogenetic comparison of the two strains HKI4363/88 and BA2968 by chromosomal single nucleotide polymorphism (SNP) analysis grouped these organisms within their relatives of the minor canonical A-branch canSNP-group A.Br.003/004 (A.Br.V770) or canonical B-branch B.Br.001/002, respectively. Strain HKI4363/88 clustered relatively closely with other members of the A.Br.003/004 lineage from Europe, South Africa, and South America. In contrast, strain BA2968 clearly constituted a new sublineage within B.Br.001/002 with its closest relative being HYO01 from South Korea.
Conclusions: Our results suggest that Finland harbors both unique (autochthonous) and more widely distributed, common clades of B. anthracis. We suspect that members of the common clades such as strains HKI4363/88 have been introduced only recently by anthropogenic activities involving importation of contaminated animal products. On the other hand, autochthonous strains such as isolate BA2968 probably have an older history of their introduction into Finland as evidenced by a high number of single nucleotide variant sites in their genomes.
Keywords: Bacillus anthracis; Comparative genomics; Finland; Multiple locus variable number of tandem repeat analysis (VNTR, MLVA); Single nucleotide polymorphism (SNP); Whole genome sequencing (WGS).
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Neglected zoonotic diseases. http://www.who.int/neglected_diseases/zoonoses/infections_more/en/. Accessed 11 July 2018.
-
- Shadomy S, El Idrissi A, Raizman E, Bruni M, Palamara E, Pittiglio C, Lubroth J. Anthrax outbreaks: a warning for heightened awareness. In: Division APaH, editor. EMPRES Watch. Rome: Food and Agricultur Organization of the United nations (FAO); 2016. p. 8.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

