The comings and goings of PARP-1 in response to DNA damage
- PMID: 30177435
- PMCID: PMC6637744
- DOI: 10.1016/j.dnarep.2018.08.022
The comings and goings of PARP-1 in response to DNA damage
Abstract
Poly(ADP-ribose) polymerase (PARP) enzymes are broadly involved in the cellular response to DNA damage. PARP-1 is the chief human PARP enzyme involved in the DNA damage response, acting as a first responder that detects DNA strand breaks, and contributes to repair pathway choice and the efficiency of repair through modulation of chromatin structure and through interaction with and modification of a multitude of DNA repair factors. This perspective summarizes our knowledge of PARP-1 involvement in DNA repair pathways, and highlights recent structural and functional data regarding the activation of PARP-1 upon detecting DNA damage, and the release and trapping of PARP-1 at sites of DNA damage.
Keywords: DNA damage response; PARP; Poly(ADP-ribose).
Copyright © 2018 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of Interest Statement
The author claims no conflict of interest.
Figures
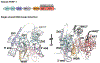
References
-
- Hassa PO, Hottiger MO, The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases., Front. Biosci. 13 (2008) 3046–82. http://www.ncbi.nlm.nih.gov/pubmed/17981777 (accessed March 22, 2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

