Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis
- PMID: 30177740
- PMCID: PMC6660648
- DOI: 10.1038/s41564-018-0230-7
Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis
Abstract
Staphylococcus epidermidis is a conspicuous member of the human microbiome, widely present on healthy skin. Here we show that S. epidermidis has also evolved to become a formidable nosocomial pathogen. Using genomics, we reveal that three multidrug-resistant, hospital-adapted lineages of S. epidermidis (two ST2 and one ST23) have emerged in recent decades and spread globally. These lineages are resistant to rifampicin through acquisition of specific rpoB mutations that have become fixed in the populations. Analysis of isolates from 96 institutions in 24 countries identified dual D471E and I527M RpoB substitutions to be the most common cause of rifampicin resistance in S. epidermidis, accounting for 86.6% of mutations. Furthermore, we reveal that the D471E and I527M combination occurs almost exclusively in isolates from the ST2 and ST23 lineages. By breaching lineage-specific DNA methylation restriction modification barriers and then performing site-specific mutagenesis, we show that these rpoB mutations not only confer rifampicin resistance, but also reduce susceptibility to the last-line glycopeptide antibiotics, vancomycin and teicoplanin. Our study has uncovered the previously unrecognized international spread of a near pan-drug-resistant opportunistic pathogen, identifiable by a rifampicin-resistant phenotype. It is possible that hospital practices, such as antibiotic monotherapy utilizing rifampicin-impregnated medical devices, have driven the evolution of this organism, once trivialized as a contaminant, towards potentially incurable infections.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
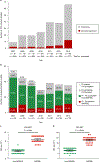



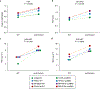

Comment in
-
Time for biocide stewardship?Nat Microbiol. 2019 May;4(5):732-733. doi: 10.1038/s41564-019-0360-6. Nat Microbiol. 2019. PMID: 30804541 No abstract available.
References
-
- Götz F Staphylococcus and biofilms. Mol. Microbiol 43, 1367–1378 (2002). - PubMed
-
- Sievert DM et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Control Hosp. Epidemiol 34, 1–14 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

