Precancerous neoplastic cells can move through the pancreatic ductal system
- PMID: 30177826
- PMCID: PMC6342205
- DOI: 10.1038/s41586-018-0481-8
Precancerous neoplastic cells can move through the pancreatic ductal system
Abstract
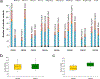
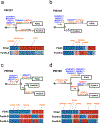
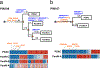
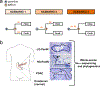
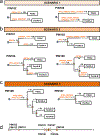
Most adult carcinomas develop from noninvasive precursor lesions, a progression that is supported by genetic analysis. However, the evolutionary and genetic relationships among co-existing lesions are unclear. Here we analysed the somatic variants of pancreatic cancers and precursor lesions sampled from distinct regions of the same pancreas. After inferring evolutionary relationships, we found that the ancestral cell had initiated and clonally expanded to form one or more lesions, and that subsequent driver gene mutations eventually led to invasive pancreatic cancer. We estimate that this multi-step progression generally spans many years. These new data reframe the step-wise progression model of pancreatic cancer by illustrating that independent, high-grade pancreatic precursor lesions observed in a single pancreas often represent a single neoplasm that has colonized the ductal system, accumulating spatial and genetic divergence over time.
Figures

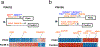




Comment in
-
Killer on the road?-cells from pancreatic preneoplastic lesions disseminate through pancreatic ducts on their way to cancer.Hepatobiliary Surg Nutr. 2019 Aug;8(4):392-394. doi: 10.21037/hbsn.2019.06.03. Hepatobiliary Surg Nutr. 2019. PMID: 31489311 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

