Single cell genomes of Prochlorococcus, Synechococcus, and sympatric microbes from diverse marine environments
- PMID: 30179231
- PMCID: PMC6122165
- DOI: 10.1038/sdata.2018.154
Single cell genomes of Prochlorococcus, Synechococcus, and sympatric microbes from diverse marine environments
Abstract
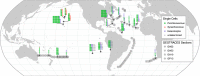
Prochlorococcus and Synechococcus are the dominant primary producers in marine ecosystems and perform a significant fraction of ocean carbon fixation. These cyanobacteria interact with a diverse microbial community that coexists with them. Comparative genomics of cultivated isolates has helped address questions regarding patterns of evolution and diversity among microbes, but the fraction that can be cultivated is miniscule compared to the diversity in the wild. To further probe the diversity of these groups and extend the utility of reference sequence databases, we report a data set of single cell genomes for 489 Prochlorococcus, 50 Synechococcus, 9 extracellular virus particles, and 190 additional microorganisms from a diverse range of bacterial, archaeal, and viral groups. Many of these uncultivated single cell genomes are derived from samples obtained on GEOTRACES cruises and at well-studied oceanographic stations, each with extensive suites of physical, chemical, and biological measurements. The genomic data reported here greatly increases the number of available Prochlorococcus genomes and will facilitate studies on evolutionary biology, microbial ecology, and biological oceanography.
Conflict of interest statement
The authors declare no competing interests.
Figures
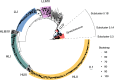

References
Data Citations
-
- Berube P. M., et al. . 2018. Figshare. https://doi.org/10.6084/m9.figshare.c.4037048 - DOI
-
- 2017. GenBank. MG666579-MG668595
-
- 2018. GenBank. MH074888-MH077527
-
- 2018. NCBI Sequence Read Archive. SRP141175
-
- 2018. GenBank. QBVH00000000-QCVZ00000000
References
-
- Karl D. M. A Sea of Change: Biogeochemical Variability in the North Pacific Subtropical Gyre. Ecosystems 2, 181–214 (1999).
-
- Partensky F. & Garczarek L. Prochlorococcus: advantages and limits of minimalism. Ann Rev Mar Sci 2, 305–331 (2010). - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

