Computational screening of known broad-spectrum antiviral small organic molecules for potential influenza HA stem inhibitors
- PMID: 30180218
- PMCID: PMC6122827
- DOI: 10.1371/journal.pone.0203148
Computational screening of known broad-spectrum antiviral small organic molecules for potential influenza HA stem inhibitors
Abstract
Background: With the emergence of new influenza virus strains that are resistant to current inhibitors such as oseltamivir (anti-neuraminidase (NA)) and amantadine (anti-M2 proton channel), influenza A viruses continue to be a serious threat to the public health worldwide. With this in view, there is a persistent need for the development of broader and more effective vaccines and therapeutics. Identification of broadly neutralizing antibodies (bNAbs) that recognize relatively invariant structures on influenza haemagglutinin (HA) stem has invigorated efforts to develop universal influenza vaccines.
Aim: The current computational study is designed to identify potential flavonoid inhibitors that bind to the contact epitopes of HA stem that are targeted by broadly neutralizing antibodies (bNAb).
Method: In this study, we utilized the three-dimensional crystallographic structure of different HA subtypes (H1, H2, H5, H3, and H7) in complex with bNAb to screen for potential broadly reactive influenza inhibitors. We performed Quantitative Structure-Activity and Relationship (QSAR) for 100 natural compounds known for their antiviral activity and performed molecular docking using AutoDock 4.2 suite. Furthermore, we conducted virtual screening of 1413 bioassay hit compounds by using virtual lab bench CLC Drug Discovery.
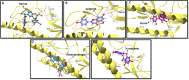
Results: The results showed 18 lead flavonoids with strong binding abilities to bNAb epitopes of various HA subtypes. These 18 broadly reactive compounds exhibited significant interactions with an average of seven Hbonds, docking energy of -22.43 kcal·mol-1, and minimum interaction energy of -4.65 kcal·mol-1, with functional contact residues. Procyanidin depicted strong interactions with group 1 HAs, whereas both sorbitol and procyanidin exhibited significant interactions with group 2 HAs.
Conclusion: Using in silico docking analysis, we identified 18 bioactive flavonoids with potential strong binding cababilities to influenza HA-stems of various subtypes, which are the target for bNAb. The virtual screened bioassay hit compounds depicted a high number of Hbonds but low interaction and docking values compared to antiviral flavonoids. Using structure-based design and nanotechnology-based approaches, identified molecules could be modified to generate next generation anti-influenza drugs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Recent progress and challenges in the discovery of new neuraminidase inhibitors.Expert Opin Ther Pat. 2013 Apr;23(4):409-23. doi: 10.1517/13543776.2013.765861. Epub 2013 Feb 1. Expert Opin Ther Pat. 2013. PMID: 23369206 Review.
-
An influenza A hemagglutinin small-molecule fusion inhibitor identified by a new high-throughput fluorescence polarization screen.Proc Natl Acad Sci U S A. 2020 Aug 4;117(31):18431-18438. doi: 10.1073/pnas.2006893117. Epub 2020 Jul 20. Proc Natl Acad Sci U S A. 2020. PMID: 32690700 Free PMC article.
-
Antiviral potential of natural compounds against influenza virus hemagglutinin.Comput Biol Chem. 2017 Dec;71:207-218. doi: 10.1016/j.compbiolchem.2017.11.001. Epub 2017 Nov 4. Comput Biol Chem. 2017. PMID: 29149637
-
A small-molecule fragment that emulates binding of receptor and broadly neutralizing antibodies to influenza A hemagglutinin.Proc Natl Acad Sci U S A. 2018 Apr 17;115(16):4240-4245. doi: 10.1073/pnas.1801999115. Epub 2018 Apr 2. Proc Natl Acad Sci U S A. 2018. PMID: 29610325 Free PMC article.
-
[Antiviral Drugs Targeting Influenza Virus Surface Proteins: A Computational Structural Biology Approach].Yakugaku Zasshi. 2015;135(9):1015-21. doi: 10.1248/yakushi.15-00175-3. Yakugaku Zasshi. 2015. PMID: 26329546 Review. Japanese.
Cited by
-
In Silico Prediction and Bioactivity Evaluation of Chemical Ingredients Against Influenza A Virus From Isatis tinctoria L.Front Pharmacol. 2021 Dec 7;12:755396. doi: 10.3389/fphar.2021.755396. eCollection 2021. Front Pharmacol. 2021. PMID: 34950027 Free PMC article.
-
An overview of viruses discovered over the last decades and drug development for the current pandemic.Eur J Pharmacol. 2021 Jan 5;890:173746. doi: 10.1016/j.ejphar.2020.173746. Epub 2020 Nov 19. Eur J Pharmacol. 2021. PMID: 33221318 Free PMC article. Review.
-
Recent progress in chemical approaches for the development of novel neuraminidase inhibitors.RSC Adv. 2021 Jan 6;11(3):1804-1840. doi: 10.1039/d0ra07283d. eCollection 2021 Jan 4. RSC Adv. 2021. PMID: 35424082 Free PMC article. Review.
-
In silico virtual screening of lead compounds for major antigenic sites in respiratory syncytial virus fusion protein.Emergent Mater. 2022;5(2):295-305. doi: 10.1007/s42247-021-00213-6. Epub 2021 May 3. Emergent Mater. 2022. PMID: 33969268 Free PMC article.
-
Debulking different Corona (SARS-CoV-2 delta, omicron, OC43) and Influenza (H1N1, H3N2) virus strains by plant viral trap proteins in chewing gums to decrease infection and transmission.Biomaterials. 2022 Sep;288:121671. doi: 10.1016/j.biomaterials.2022.121671. Epub 2022 Jul 18. Biomaterials. 2022. PMID: 35953331 Free PMC article. Clinical Trial.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

