Systematic pharmacological screens uncover novel pathways involved in cerebral cavernous malformations
- PMID: 30181117
- PMCID: PMC6180302
- DOI: 10.15252/emmm.201809155
Systematic pharmacological screens uncover novel pathways involved in cerebral cavernous malformations
Abstract
Cerebral cavernous malformations (CCMs) are vascular lesions in the central nervous system causing strokes and seizures which currently can only be treated through neurosurgery. The disease arises through changes in the regulatory networks of endothelial cells that must be comprehensively understood to develop alternative, non-invasive pharmacological therapies. Here, we present the results of several unbiased small-molecule suppression screens in which we applied a total of 5,268 unique substances to CCM mutant worm, zebrafish, mouse, or human endothelial cells. We used a systems biology-based target prediction tool to integrate the results with the whole-transcriptome profile of zebrafish CCM2 mutants, revealing signaling pathways relevant to the disease and potential targets for small-molecule-based therapies. We found indirubin-3-monoxime to alleviate the lesion burden in murine preclinical models of CCM2 and CCM3 and suppress the loss-of-CCM phenotypes in human endothelial cells. Our multi-organism-based approach reveals new components of the CCM regulatory network and foreshadows novel small-molecule-based therapeutic applications for suppressing this devastating disease in patients.
Keywords: CCM; ERK5; KLF2; angiogenesis; indirubin‐3‐monoxime.
© 2018 The Authors. Published under the terms of the CC BY 4.0 license.
Figures

- A
Overview of the four different screening assays used in this study. Zebrafish embryos and C. elegans are screened in 24‐well and 96‐well plates, respectively. The most promising active compounds are retested in shCCM2 HUVECs. One compound is tested for suppression of vascular lesion formation in the cerebellum of iCCM2 and iCCM3 mouse models.
- B
Overlap of rescue compounds screened in the different assays.
- C–E
Examples of rescue of cardiovascular defects of the zebrafish ccm2 m201 mutant. Inverted images of confocal z‐scan projections of the 46 hpf head region and heart (endocardium) of wild‐type (WT) and ccm2 m201 mutant zebrafish embryos carrying the endothelial Tg(kdrl:GFP)s843 reporter transgene. Embryos are untreated (C) or treated between 17 and 48 hpf with 10 μM of the Lck inhibitor C8863 (D) or with 10 μM of the ERK5 inhibitor XMD8‐92 (E). Both compounds resulted in a reduction in heart size and narrowing of the heart tube at the atrioventricular canal (arrowheads). Scale bar is 100 μm.

- A
kri‐1 mutant worms treated with control L4440 RNAi and with DMSO are viable. Shown is a representative picture of a control well from a 96‐well plate.
- B
Treatment of kri‐1 mutants with ccm‐3 RNAi causes synthetic lethality. Incubation of this strain with DMSO (control) has no further effect on the synthetic lethality.
- C
Incubation of the ccm‐3 RNAi‐treated kri‐1 mutants with the phospholipase C inhibitor ET‐18‐OCH3 results in a mild rescue, as seen by the presence of a few worms.
- D
Incubation of the ccm‐3 RNAi‐treated kri‐1 mutants with the GSK‐3/PI3K/Akt/mTOR inhibitor TWS119 strongly rescues synthetic as indicated by the high number of worms.
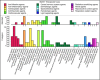

- A
MeSH terms for physiological effects.
- B
MeSH terms for molecular mechanisms.


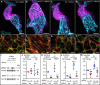
- A–D
Treatment with 5 μM indirubin‐3‐monoxime (IR3mo) rescues the embryonic zebrafish ccm2 m201 mutant ballooning heart phenotype. Shown are images of confocal z‐scan projections of wild‐type (WT) (A, C) and ccm2 m201 mutant (B, D) zebrafish embryonic hearts at 48 hpf expressing the endothelial reporter Tg(kdrl:GFP)s843 (cyan) and counter‐stained for Alcam (magenta, labeling the myocardium). Scale bar is 50 μm.
- E–H
Treatment with 10 μM IR3mo for 48 h restores the wild‐type ACTIN‐adhesive phenotype in KRIT1‐depleted HUVECs. Shown are confocal images of HUVECs with labeled VE‐cadherin (green) and F‐ACTIN (red). Control siRNA‐silenced (E, G) and KRIT1 siRNA‐silenced (F, H) HUVECs were not treated (E, F) or treated (G, H) with IR3mo. Scale bar is 10 μm.
- I–K
Treatment with IR3mo reduces phosphorylation of ERK (pERK) without affecting overall ERK protein levels. Shown in (I) are representative Western blots of KRIT1‐silenced HUVECs lysates treated or not with 10 μM of IR3mo during transfection. ERK5 (J) and phosphorylated ERK5 protein levels (K) were measured relative to ACTIN protein levels based on three biological replicates with two technical replicates each (n = 3). Statistical analyses were performed using two‐way ANOVA followed by Tukey's multiple comparisons test; error bars are SD. ****P < 0.0001; ns, not significant.
- L
Treatment with IR3mo rescues the elevated KLF2 levels of KRIT1‐silenced HUVECs. RT‐qPCRs were performed to measure KLF2 levels, on three biological replicates (n = 3). Statistical analyses were performed using one‐way ANOVA followed by Tukey's multiple comparisons test; error bars are SD. ***P < 0.001; ns, not significant.
- M
Treatment with IR3mo rescues the elevated klf2a levels of ccm2 m201 mutant zebrafish embryos at 48 hpf. RT‐qPCRs were performed to measure klf2a levels, on six biological replicates (n = 6). Statistical analyses were performed using one‐way ANOVA followed by Tukey's multiple comparisons test; error bars are SD. *P < 0.05; ns, not significant.

- A–D
Treatment with 5 μM IR3mo rescues the embryonic zebrafish krit1 ty219c mutant ballooning heart phenotype. Shown are images of confocal z‐scan projections of wild‐type (WT) (A, C) and krit1 ty219 mutant (B, D) zebrafish embryonic hearts at 48 hpf expressing the endothelial reporter Tg(kdrl:GFP)s843 (green) and counter‐stained for ACTIN (red). Scale bar is 50 μm.
- E–J
Treatment with 10 μM IR3mo for 48 h rescued the CCM phenotype in CCM2‐ or CCM3‐depleted HUVECs. Shown are confocal images of HUVECs with labeled VE‐cadherin (green) and F‐ACTIN (red). Control siRNA‐silenced (E, F), CCM2 siRNA‐silenced (G, H), or CCM3 siRNA‐silenced (I, J) HUVECs untreated (E, G, I) or treated (F, H, J) with IR3mo. Scale bar is 10 μm.

- A–D
Shown are representative pictures of brains at P8 from iCCM2 (A, B) or iCCM3 (C, D) mice treated with vehicle (A, C) or with IR3mo (B, D). Mice were treated daily between P2 and P7 with vehicle or with IR3mo. Scale bar is 1 mm.
- E
Treatment with IR3mo does not affect cerebellum width at P8 of iCCM2 mice. The measurements were performed on seven vehicle‐treated and eight IR3mo‐treated mice (n = 7, n = 8). Student's two‐tailed t‐tests were performed; error bars are SD. ns, not significant.
- F
Treatment with IR3mo does not affect animal weight at P8 of iCCM2 mice. The measurements were performed on eight vehicle‐treated and eight IR3mo‐treated mice (n = 8, n = 8). Student's two‐tailed t‐tests were performed; error bars are SD. ns, not significant.
- G, H
Quantifications of relative numbers of caverns within three categories based on cavern size. Cerebellar regions were measured in iCCM2 and iCCM3 mutants at P8 after treatment with vehicle (n = 8 for iCCM2 and n = 5 for iCCM3) or IR3mo (n = 8 for iCCM2 and n = 6 for iCCM3). Student's two‐tailed t‐tests were performed; error bars are SD. ns, not significant; *P < 0.05; **P < 0.01.
- I
Model scheme showing the molecular effect of IR3mo treatment on the ERK5 phosphorylation levels and the KLF2 expression levels in the CCM loss‐of‐function context. Two arrowheads indicate different molecular effects of IR3mo within the ERK5‐KLF2 pathway.
References
-
- Berthold MR, Cebron N, Dill F, Gabriel TR, Kötter T, Meinl T, Ohl P, Sieb C, Thiel K, Wiswedel B (2007) KNIME: the Konstanz information miner In Studies in classification, data analysis, and knowledge organization (GfKL 2007). Springer‐Verlag Berlin Heidelberg.
-
- Boulday G, Blécon A, Petit N, Chareyre F, Garcia LA, Niwa‐Kawakita M, Giovannini M, Tournier‐Lasserve E (2009) Tissue‐specific conditional CCM2 knockout mice establish the essential role of endothelial CCM2 in angiogenesis: implications for human cerebral cavernous malformations. Dis Model Mech 2: 168–177 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

