Interplay of ancestral non-primate lentiviruses with the virus-restricting SAMHD1 proteins of their hosts
- PMID: 30181218
- PMCID: PMC6200947
- DOI: 10.1074/jbc.RA118.004567
Interplay of ancestral non-primate lentiviruses with the virus-restricting SAMHD1 proteins of their hosts
Abstract
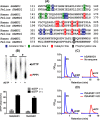
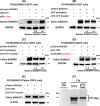
Lentiviruses infect both dividing CD4+ T cells and nondividing myeloid cells, and the infected myeloid cells serve as long-living viral reservoirs. Host sterile alpha motif- and histidine-aspartate domain-containing protein 1 (SAMHD1) kinetically restricts reverse transcription of primate lentiviruses, including human immunodeficiency virus, type 1 (HIV-1) and simian immunodeficiency virus (SIV), in nondividing myeloid cells. SAMHD1 enforces this restriction through its dNTP triphosphohydrolase (dNTPase) activity that depletes cellular dNTPs. Some primate lentiviruses, such as HIV-2, SIVsm, and SIVagm, counteract SAMHD1 restriction by using their viral accessory proteins (Vpx or Vpr) that induce the proteosomal degradation of SAMHD1 and increase dNTP levels. SAMHD1 is conserved among non-primate mammals such as cats, cows, and horses that also carry their own lentiviruses (feline and bovine immunodeficiency viruses and equine infectious anemia viruses, respectively). However, whether these viruses also target SAMHD1 is unknown. Here, we tested whether these ancestral non-primate lentiviruses also can counteract their host SAMHD1 proteins by promoting their proteosomal degradation. Using biochemical and various cell-based assays, we observed that SAMHD1 proteins from the non-primate host species display dGTP-dependent dNTPase activity, but that the non-primate lentiviruses fail to proteosomally degrade the SAMHD1 proteins of their hosts. Our findings suggest that accessory protein-mediated proteosomal degradation of SAMHD1 did not exist among the ancestral non-primate lentiviruses and was uniquely gained by some primate lentiviruses after their transmission to primate species.
Keywords: SAM domain and HD domain-containing protein 1 (SAMHD1); antiviral defense; dNTPase; immunodeficiency virus; lentivirus; macrophage; nucleotide; proteosomal degradation; reverse transcription.
© 2018 Mereby et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

