Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions
- PMID: 30181241
- PMCID: PMC6130803
- DOI: 10.1126/scisignal.aat6409
Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions
Abstract
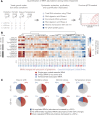
Decoding the information in mRNA during protein synthesis relies on tRNA adaptors, the abundance of which can affect the decoding rate and translation efficiency. To determine whether cells alter tRNA abundance to selectively regulate protein expression, we quantified changes in the abundance of individual tRNAs at different time points in response to diverse stress conditions in Saccharomyces cerevisiae We found that the tRNA pool was dynamic and rearranged in a manner that facilitated selective translation of stress-related transcripts. Through genomic analysis of multiple data sets, stochastic simulations, and experiments with designed sequences of proteins with identical amino acids but altered codon usage, we showed that changes in tRNA abundance affected protein expression independently of factors such as mRNA abundance. We suggest that cells alter their tRNA abundance to selectively affect the translation rates of specific transcripts to increase the amounts of required proteins under diverse stress conditions.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures




Comment in
-
Coping with stress by regulating tRNAs.Sci Signal. 2018 Sep 4;11(546):eaau1098. doi: 10.1126/scisignal.aau1098. Sci Signal. 2018. PMID: 30181239
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

