Polyamine metabolism and cancer: treatments, challenges and opportunities
- PMID: 30181570
- PMCID: PMC6487480
- DOI: 10.1038/s41568-018-0050-3
Polyamine metabolism and cancer: treatments, challenges and opportunities
Abstract
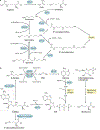
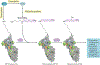
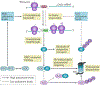
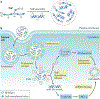
Advances in our understanding of the metabolism and molecular functions of polyamines and their alterations in cancer have led to resurgence in the interest of targeting polyamine metabolism as an anticancer strategy. Increasing knowledge of the interplay between polyamine metabolism and other cancer-driving pathways, including the PTEN-PI3K-mTOR complex 1 (mTORC1), WNT signalling and RAS pathways, suggests potential combination therapies that will have considerable clinical promise. Additionally, an expanding number of promising clinical trials with agents targeting polyamines for both therapy and prevention are ongoing. New insights into molecular mechanisms linking dysregulated polyamine catabolism and carcinogenesis suggest additional strategies that can be used for cancer prevention in at-risk individuals. In addition, polyamine blocking therapy, a strategy that combines the inhibition of polyamine biosynthesis with the simultaneous blockade of polyamine transport, can be more effective than therapies based on polyamine depletion alone and may involve an antitumour immune response. These findings open up new avenues of research into exploiting aberrant polyamine metabolism for anticancer therapy.
Conflict of interest statement
Competing interests
The authors receive US National Institutes of Health grant support (R.A.C.) and are patent holders (R.A.C. and A.E.P.).T.M.S. declares no competing interests.
Figures

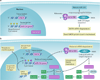
References
-
- Terui Y et al. Polyamines protect nucleic acids against depurination. Int. J. Biochem. Cell Biol 99, 147–153 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

