Transcriptome sequencing reveals key potential long non-coding RNAs related to duration of fertility trait in the uterovaginal junction of egg-laying hens
- PMID: 30181614
- PMCID: PMC6123486
- DOI: 10.1038/s41598-018-31301-z
Transcriptome sequencing reveals key potential long non-coding RNAs related to duration of fertility trait in the uterovaginal junction of egg-laying hens
Abstract
Duration of fertility, (DF) is an important functional trait in poultry production and lncRNAs have emerged as important regulators of various process including fertility. In this study we applied a genome-guided strategy to reconstruct the uterovaginal junction (UVJ) transcriptome of 14 egg-laying birds with long- and short-DF (n = 7); and sought to uncover key lncRNAs related to duration of fertility traits by RNA-sequencing technology. Examination of RNA-seq data revealed a total of 9977 lncRNAs including 2576 novel lncRNAs. Differential expression (DE) analysis of lncRNA identified 223 lncRNAs differentially expressed between the two groups. DE-lncRNA target genes prediction uncovered over 200 lncRNA target genes and functional enrichment tests predict a potential function of DE-lncRNAs. Gene ontology classification and pathway analysis revealed 8 DE-lncRNAs, with the majority of their target genes enriched in biological functions such as reproductive structure development, developmental process involved in reproduction, response to cytokine, carbohydrate binding, chromatin organization, and immune pathways. Differential expression of lncRNAs and target genes were confirmed by qPCR. Together, these results significantly expand the utility of the UVJ transcriptome and our analysis identification of key lncRNAs and their target genes regulating DF will form the baseline for understanding the molecular functions of lncRNAs regulating DF.
Conflict of interest statement
The authors declare no competing interests.
Figures
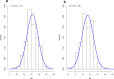



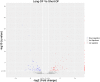



References
-
- Brillard J. Practical aspects of fertility in poultry. World’s Poultry Science Journal. 2003;59:441–446. doi: 10.1079/WPS20030027. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

