Identification, expression and characterization of the recombinant Sol g 4.1 protein from the venom of the tropical fire ant Solenopsis geminata
- PMID: 30181738
- PMCID: PMC6116302
- DOI: 10.1186/s40409-018-0159-6
Identification, expression and characterization of the recombinant Sol g 4.1 protein from the venom of the tropical fire ant Solenopsis geminata
Abstract
Background: Fire ant venom is a complex mixture consisting of basic piperidine alkaloids, various biologically active peptides and protein components, including a variety of major allergenic proteins. Tropical fire ant Solenopsis geminata is an important stinging ant species that causes anaphylaxis and serious medical problems. Although the biological activities of allergenic venom proteins that are unique to ant venom, particularly Solenopsis 2 and 4, are still unknown, these proteins are believed to play important roles in mediating the effects of the piperidine derivatives in the venom.
Methods: In the present study, the cDNA cloning, sequencing and three-dimensional structure of Sol g 4.1 venom protein are described. The recombinant Sol g 4.1 protein (rSol g 4.1) was produced in E. coli, and its possible function as a hydrophobic binding protein was characterized by paralyzing crickets using the 50% piperidine dose (PD50). Moreover, an antiserum was produced in mice to determine the allergenic properties of Sol g 4.1, and the antiserum was capable of binding to Sol g 4.1, as determined by Western blotting.
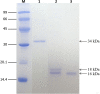
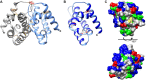
Results: The molecular weight of Sol g 4.1 protein is 16 kDa, as determined by SDS-PAGE. The complete cDNA is 414 bp in length and contains a leader sequence of 19 amino acids. The protein consists of six cysteines that presumably form three disulfide bonds, based on a predicted three-dimensional model, creating the interior hydrophobic pocket and stabilizing the structure. The rSol g 4.1 protein was expressed in inclusion bodies, as determined by SDS-PAGE. Dialysis techniques were used to refold the recombinant protein into the native form. Its secondary structure, which primarily consists of α-helices, was confirmed by circular dichroism analysis, and the three-dimensional model was also verified. The results of allergenic analysis performed on mice showed that the obtained protein was predicted to be allergenically active. Moreover, we report on the possible role of the Sol g 4.1 venom protein, which significantly reduced the PD50 from 0.027 to 0.013% in paralyzed crickets via synergistic effects after interactions with piperidine alkaloids.
Conclusions: The primary structure of Sol g 4.1 showed high similarity to that of venom proteins in the Solenopsis 2 and 4 family. Those proteins are life-threatening and produce IgE-mediated anaphylactic reactions in allergic individuals. The possible function of this protein is the binding of the interior hydrophobic pockets with piperidine alkaloids, as determined by the analysis of the structural model and PD50 test.
Keywords: Allergen; Fire ant; Sol g 4.1 protein; Stinging ant; Venom protein.
Conflict of interest statement
The present study was approved by the Animal Ethics Committee of Khon Kaen University based on the Ethics for Animal Experimentation of the National Research Council of Thailand (reference no. 0514.1.12.2/66).Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


 ). Alignment of the six cysteines (red stars) between all Solenopsis 2 and 4 genes and the alignment of the seventh cysteine in the Sol 2 genes (pink star). Residues lining the interior face of the Sol g 4.1 protein are indicated by x. The sequences were submitted to GenBank with the following accession numbers: Solenopsis 2 proteins: P35775 for Sol i 2, P35776 for Sol r 2, ABC58726 for Sol s 2, ALM98859 for Sol × 2, AAY32928 for Sol i 2q and AAY32926 for Sol g 2q; and Solenopsis 4 proteins: AAC97369 for Sol i 4.01, AAC97370 for Sol i 4.02, AAF65312 for Sol g 4.01, AAF65313 for Sol g 4.02, AAY32927 for Sol g 4q and AAY32929 for Sol i 4q
). Alignment of the six cysteines (red stars) between all Solenopsis 2 and 4 genes and the alignment of the seventh cysteine in the Sol 2 genes (pink star). Residues lining the interior face of the Sol g 4.1 protein are indicated by x. The sequences were submitted to GenBank with the following accession numbers: Solenopsis 2 proteins: P35775 for Sol i 2, P35776 for Sol r 2, ABC58726 for Sol s 2, ALM98859 for Sol × 2, AAY32928 for Sol i 2q and AAY32926 for Sol g 2q; and Solenopsis 4 proteins: AAC97369 for Sol i 4.01, AAC97370 for Sol i 4.02, AAF65312 for Sol g 4.01, AAF65313 for Sol g 4.02, AAY32927 for Sol g 4q and AAY32929 for Sol i 4q


Similar articles
-
Characterization and Localization of Sol g 2.1 Protein from Solenopsis geminata Fire Ant Venom in the Central Nervous System of Injected Crickets (Acheta domestica).Int J Mol Sci. 2023 Oct 1;24(19):14814. doi: 10.3390/ijms241914814. Int J Mol Sci. 2023. PMID: 37834262 Free PMC article.
-
Protein-Ligand Binding and Structural Modelling Studies of Pheromone-Binding Protein-like Sol g 2.1 from Solenopsis geminata Fire Ant Venom.Molecules. 2024 Feb 27;29(5):1033. doi: 10.3390/molecules29051033. Molecules. 2024. PMID: 38474545 Free PMC article.
-
Structure-based epitope prediction and assessment of cross-reactivity of Myrmecia pilosula venom-specific IgE and recombinant Sol g proteins (Solenopsis geminata).Sci Rep. 2024 May 15;14(1):11145. doi: 10.1038/s41598-024-61843-4. Sci Rep. 2024. PMID: 38750087 Free PMC article.
-
Ant allergens and hypersensitivity reactions in response to ant stings.Asian Pac J Allergy Immunol. 2015 Dec;33(4):267-75. Asian Pac J Allergy Immunol. 2015. PMID: 26708389 Review.
-
Hypersensitivity to fire ant venom.Ann Allergy Asthma Immunol. 1996 Aug;77(2):87-95; quiz 96-9. doi: 10.1016/S1081-1206(10)63493-X. Ann Allergy Asthma Immunol. 1996. PMID: 8760773 Review.
Cited by
-
Unique venom proteins from Solenopsis invicta x Solenopsis richteri hybrid fire ants.Toxicon X. 2021 May 7;9-10:100065. doi: 10.1016/j.toxcx.2021.100065. eCollection 2021 Jul. Toxicon X. 2021. PMID: 34027387 Free PMC article.
-
Heterologous expression and mutagenesis of recombinant Vespa affinis hyaluronidase protein (rVesA2).J Venom Anim Toxins Incl Trop Dis. 2019 Dec 5;25:e20190030. doi: 10.1590/1678-9199-JVATITD-2019-0030. eCollection 2019. J Venom Anim Toxins Incl Trop Dis. 2019. PMID: 31839801 Free PMC article.
-
Characterization and Localization of Sol g 2.1 Protein from Solenopsis geminata Fire Ant Venom in the Central Nervous System of Injected Crickets (Acheta domestica).Int J Mol Sci. 2023 Oct 1;24(19):14814. doi: 10.3390/ijms241914814. Int J Mol Sci. 2023. PMID: 37834262 Free PMC article.
-
Biological Activities and Ecological Significance of Fire Ant Venom Alkaloids.Toxins (Basel). 2023 Jul 3;15(7):439. doi: 10.3390/toxins15070439. Toxins (Basel). 2023. PMID: 37505709 Free PMC article. Review.
-
The Peptide Venom Composition of the Fierce Stinging Ant Tetraponera aethiops (Formicidae: Pseudomyrmecinae).Toxins (Basel). 2019 Dec 14;11(12):732. doi: 10.3390/toxins11120732. Toxins (Basel). 2019. PMID: 31847368 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

