Protocol: a versatile, inexpensive, high-throughput plant genomic DNA extraction method suitable for genotyping-by-sequencing
- PMID: 30181764
- PMCID: PMC6114050
- DOI: 10.1186/s13007-018-0336-1
Protocol: a versatile, inexpensive, high-throughput plant genomic DNA extraction method suitable for genotyping-by-sequencing
Abstract
Background: The recent development of next-generation sequencing DNA marker technologies, such as genotyping-by-sequencing (GBS), generates thousands of informative single nucleotide polymorphism markers in almost any species, regardless of genomic resources. This enables poorly resourced or "orphan" crops/species access to high-density, high-throughput marker platforms which have revolutionised population genetics studies and plant breeding. DNA quality underpins success of GBS methods as the DNA must be amenable to restriction enzyme digestion and sequencing. A barrier to implementing GBS technologies is access to inexpensive, high-throughput extraction methods that yield sequencing-quality genomic DNA (gDNA) from plants. Several high-throughput DNA extraction methods are available, but typically provide low yield or poor quality gDNA, or are costly (US$6-$9/sample) for consumables.
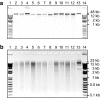
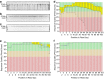
Results: We modified a non-organic solvent protocol to extract microgram quantities (1-13 μg) of sequencing-quality high molecular weight gDNA inexpensively in 96-well plates from either fresh, freeze-dried or silica gel-dried plant tissue. The protocol was effective for several easy and difficult-to-extract forage, crop, horticultural and common model species including Trifolium, Medicago, Lolium, Secale, Festuca, Malus, Oryza, and Arabidopsis. The extracted DNA was of high molecular weight and digested readily with restriction enzymes. Contrasting with other extraction protocols we assessed, Illumina-based sequencing of GBS libraries developed from this gDNA had very uniform high quality base-calls to the end of sequence reads. Furthermore, DNA extracted using this method has been sequenced successfully with the PacBio long-read platform. The protocol is scalable, readily automated without requirement for fume hoods, requires approximately three hours to process 192 samples (384-576 samples/day), and is inexpensive at US$0.62/sample for consumables.
Conclusions: This versatile, scalable and simple protocol yields high molecular weight genomic DNA suitable for restriction enzyme digestion and next-generation sequencing applications including GBS and long-read sequencing platforms such as PacBio. The low cost, high-throughput, and extraction of high quality gDNA from a range of fresh and dried source plant material makes this method suitable for many sequencing and genotyping applications including large-scale sample screening underpinning breeding programmes.
Keywords: Arabidopsis; Festuca; Freeze-dried; High-throughput; Lolium; Malus; Medicago; Next-generation sequencing; Oryza; Secale; Silica gel-dried; Trifolium.
Figures


References
-
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15.
LinkOut - more resources
Full Text Sources
Other Literature Sources

