Draft genome of Dugesia japonica provides insights into conserved regulatory elements of the brain restriction gene nou-darake in planarians
- PMID: 30181897
- PMCID: PMC6114478
- DOI: 10.1186/s40851-018-0102-2
Draft genome of Dugesia japonica provides insights into conserved regulatory elements of the brain restriction gene nou-darake in planarians
Abstract
Background: Planarians are non-parasitic Platyhelminthes (flatworms) famous for their regeneration ability and for having a well-organized brain. Dugesia japonica is a typical planarian species that is widely distributed in the East Asia. Extensive cellular and molecular experimental methods have been developed to identify the functions of thousands of genes in this species, making this planarian a good experimental model for regeneration biology and neurobiology. However, no genome-level information is available for D. japonica, and few gene regulatory networks have been identified thus far.
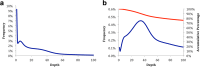
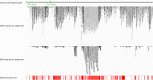
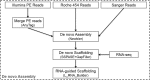
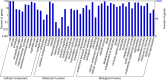
Results: To obtain whole-genome information on this species and to study its gene regulatory networks, we extracted genomic DNA from 200 planarians derived from a laboratory-bred asexual clonal strain, and sequenced 476 Gb of data by second-generation sequencing. Kmer frequency graphing and fosmid sequence analysis indicated a complex genome that would be difficult to assemble using second-generation sequencing short reads. To address this challenge, we developed a new assembly strategy and improved the de novo genome assembly, producing a 1.56 Gb genome sequence (DjGenome ver1.0, including 202,925 scaffolds and N50 length 27,741 bp) that covers 99.4% of all 19,543 genes in the assembled transcriptome, although the genome is fragmented as 80% of the genome consists of repeated sequences (genomic frequency ≥ 2). By genome comparison between two planarian genera, we identified conserved non-coding elements (CNEs), which are indicative of gene regulatory elements. Transgenic experiments using Xenopus laevis indicated that one of the CNEs in the Djndk gene may be a regulatory element, suggesting that the regulation of the ndk gene and the brain formation mechanism may be conserved between vertebrates and invertebrates.
Conclusion: This draft genome and CNE analysis will contribute to resolving gene regulatory networks in planarians. The genome database is available at: http://www.planarian.jp.
Keywords: Conserved non-coding elements; Dugesia japonica; Genome; Nou-darake; Planarian.
Conflict of interest statement
Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Pallas PS. Spicilegia zoologica quibus novae imprimis et obscurae animaliu speciosiconibus atque conamentariis illustratur. Fasc X, Berolini. 1774.
-
- Kawakatsu M, Oki I, Tamura S. Taxonomy and geographical-distribution of Dugesia-japonica and D-Ryukyuensis in the far-east. Hydrobiologia. 1995;305:55–61. doi: 10.1007/BF00036363. - DOI
-
- Pagán OR. The first brain: the neuroscience of planarians. 1st edition: Oxford University Press; 2014.
LinkOut - more resources
Full Text Sources
Other Literature Sources

