Multiple Histone Methyl-Lysine Readers Ensure Robust Development and Germline Immortality in Caenorhabditis elegans
- PMID: 30185429
- PMCID: PMC6218232
- DOI: 10.1534/genetics.118.301518
Multiple Histone Methyl-Lysine Readers Ensure Robust Development and Germline Immortality in Caenorhabditis elegans
Abstract
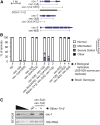
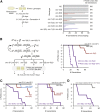
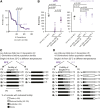
Chromatin modifications, including methylation of histone H3 at lysine 27 (H3K27me) by the Polycomb group proteins, play a broadly conserved role in the maintenance of cell fate. Diverse chromatin organization modifier (chromo) domain proteins act as "readers" of histone methylation states. However, understanding the functional relationships among chromo domains and their roles in the inheritance of gene expression patterns remains challenging. Here, we identify two chromo-domain proteins, CEC-1 and CEC-6, as potential readers of H3K27me in Caenorhabditis elegans, where they have divergent expression patterns and contribute to distinct phenotypes. Both cec-1 and cec-6 genetically interact with another chromo-domain gene, cec-3, a reader of H3K9 methylation. Combined loss of cec-1 and cec-3 leads to developmental defects in the adult that result in decreased fitness. Furthermore, loss of cec-6 and cec-3 surprisingly leads to a progressive loss of fertility across generations, a "mortal germline" phenotype. Our results provide evidence of functional compensation between H3K27me and H3K9me heterochromatin pathways, and show that histone methylation readers contribute to both somatic development and transgenerational fitness.
Keywords: C. elegans; CEC; H3K27me3; PRC2; Polycomb proteins; chromodomain proteins; gene silencing; histone methyl-lysine readers; mortal germline; worm.
Copyright © 2018 by the Genetics Society of America.
Figures





Similar articles
-
Emerging Roles for Chromo Domain Proteins in Genome Organization and Cell Fate in C. elegans.Front Cell Dev Biol. 2020 Oct 23;8:590195. doi: 10.3389/fcell.2020.590195. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33195254 Free PMC article. Review.
-
Histone H3K9 methylation promotes formation of genome compartments in Caenorhabditis elegans via chromosome compaction and perinuclear anchoring.Proc Natl Acad Sci U S A. 2020 May 26;117(21):11459-11470. doi: 10.1073/pnas.2002068117. Epub 2020 May 8. Proc Natl Acad Sci U S A. 2020. PMID: 32385148 Free PMC article.
-
Active chromatin marks drive spatial sequestration of heterochromatin in C. elegans nuclei.Nature. 2019 May;569(7758):734-739. doi: 10.1038/s41586-019-1243-y. Epub 2019 May 22. Nature. 2019. PMID: 31118512
-
MORC-1 Integrates Nuclear RNAi and Transgenerational Chromatin Architecture to Promote Germline Immortality.Dev Cell. 2017 May 22;41(4):408-423.e7. doi: 10.1016/j.devcel.2017.04.023. Dev Cell. 2017. PMID: 28535375 Free PMC article.
-
Repressive Chromatin in Caenorhabditis elegans: Establishment, Composition, and Function.Genetics. 2018 Feb;208(2):491-511. doi: 10.1534/genetics.117.300386. Genetics. 2018. PMID: 29378810 Free PMC article. Review.
Cited by
-
Genome-wide association and environmental suppression of the mortal germline phenotype of wild C. elegans.EMBO Rep. 2023 Dec 6;24(12):e58116. doi: 10.15252/embr.202358116. Epub 2023 Nov 20. EMBO Rep. 2023. PMID: 37983674 Free PMC article.
-
Systematic characterization of chromodomain proteins reveals an H3K9me1/2 reader regulating aging in C. elegans.Nat Commun. 2023 Mar 6;14(1):1254. doi: 10.1038/s41467-023-36898-y. Nat Commun. 2023. PMID: 36878913 Free PMC article.
-
Emerging Roles for Chromo Domain Proteins in Genome Organization and Cell Fate in C. elegans.Front Cell Dev Biol. 2020 Oct 23;8:590195. doi: 10.3389/fcell.2020.590195. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33195254 Free PMC article. Review.
-
SUMO-mediated regulation of H3K4me3 reader SET-26 controls germline development in C. elegans.PLoS Biol. 2025 Jan 6;23(1):e3002980. doi: 10.1371/journal.pbio.3002980. eCollection 2025 Jan. PLoS Biol. 2025. PMID: 39761316 Free PMC article.
-
The global diversity of Haemonchus contortus is shaped by human intervention and climate.Nat Commun. 2019 Oct 22;10(1):4811. doi: 10.1038/s41467-019-12695-4. Nat Commun. 2019. PMID: 31641125 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

