EGR1-Mediated Transcription of lncRNA-HNF1A-AS1 Promotes Cell-Cycle Progression in Gastric Cancer
- PMID: 30185552
- PMCID: PMC6191331
- DOI: 10.1158/0008-5472.CAN-18-1011
EGR1-Mediated Transcription of lncRNA-HNF1A-AS1 Promotes Cell-Cycle Progression in Gastric Cancer
Abstract
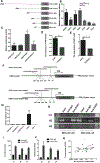
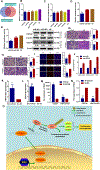
Long noncoding RNAs (lncRNA) are dysregulated in various human cancers and control tumor development and progression. However, the upstream mechanisms underlying their dysregulation remain unclear. Here, we demonstrate that the expression of hepatocyte nuclear factor 1 homeobox A antisense RNA 1 (HNF1A-AS1) is significantly upregulated in gastric cancer tissues. Overexpression of HNF1A-AS1 enhanced cell proliferation and promoted cell-cycle progression, whereas knockdown of HNF1A-AS1 elicited the opposite effects. Early growth response protein 1 (EGR1) directly bound the HNF1A-AS1 promoter region and activated its transcription. Overexpression of EGR1 enhanced cell proliferation and promoted cell-cycle promotion, similar to the function of HNF1A-AS1. HNF1A-AS1 functioned as competing endogenous RNA (ceRNA) by binding to miR-661, upregulating the expression of cell division cycle 34 (CDC34), which is a direct target of miR-661. EGR1 and HNF1A-AS1 enhanced the expression of cyclin-dependent kinase 2 (CDK2), CDK4, and cyclin E1 but inhibited the expression of p21 by promoting CDC34-mediated ubiquitination and degradation of p21. Taken together, these findings suggest that EGR1-activated HNF1A-AS1 regulates various pro- and antigrowth factors to promote the development of gastric cancer, implicating it as a possible target for therapeutic intervention in this disease.Significance: This study provides novel insights into mechanisms by which the noncoding RNA HNF1A-AS1 contributes to gastric cancer progression through modulation of the cell cycle. Cancer Res; 78(20); 5877-90. ©2018 AACR.
©2018 American Association for Cancer Research.
Conflict of interest statement
Competing financial interests
All authors promise that there is no conflict to disclose.
Figures





References
-
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87–108. - PubMed
-
- McLean MH, El-Omar EM. Genetics of gastric cancer. Nature reviews Gastroenterology & hepatology. 2014;11:664–74. - PubMed
-
- Schipper DL, Wagenmans MJ, Peters WH, Wagener DJ. Significance of cell proliferation measurement in gastric cancer. European journal of cancer. 1998;34:781–90. - PubMed
-
- Lansdorp-Vogelaar I, Kuipers EJ. Screening for gastric cancer in Western countries. Gut. 2016;65:543–4. - PubMed
-
- Duraes C, Almeida GM, Seruca R, Oliveira C, Carneiro F. Biomarkers for gastric cancer: prognostic, predictive or targets of therapy? Virchows Archiv : an international journal of pathology. 2014;464:367–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

