Structure of the membrane proximal external region of HIV-1 envelope glycoprotein
- PMID: 30185554
- PMCID: PMC6156635
- DOI: 10.1073/pnas.1807259115
Structure of the membrane proximal external region of HIV-1 envelope glycoprotein
Abstract
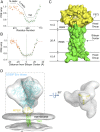
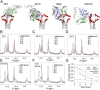
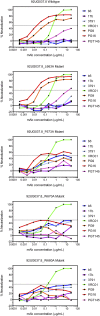
The membrane-proximal external region (MPER) of the HIV-1 envelope glycoprotein (Env) bears epitopes of broadly neutralizing antibodies (bnAbs) from infected individuals; it is thus a potential vaccine target. We report an NMR structure of the MPER and its adjacent transmembrane domain in bicelles that mimic a lipid-bilayer membrane. The MPER lies largely outside the lipid bilayer. It folds into a threefold cluster, stabilized mainly by conserved hydrophobic residues and potentially by interaction with phospholipid headgroups. Antigenic analysis and comparison with published images from electron cryotomography of HIV-1 Env on the virion surface suggest that the structure may represent a prefusion conformation of the MPER, distinct from the fusion-intermediate state targeted by several well-studied bnAbs. Very slow bnAb binding indicates that infrequent fluctuations of the MPER structure give these antibodies occasional access to alternative conformations of MPER epitopes. Mutations in the MPER not only impede membrane fusion but also influence presentation of bnAb epitopes in other regions. These results suggest strategies for developing MPER-based vaccine candidates.
Keywords: HIV-1 Env; NMR structure; membrane proximal region; transmembrane region.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. - PubMed
-
- Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. - PubMed
-
- Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

