Phylogenetic approach to recover integration dates of latent HIV sequences within-host
- PMID: 30185556
- PMCID: PMC6156657
- DOI: 10.1073/pnas.1802028115
Phylogenetic approach to recover integration dates of latent HIV sequences within-host
Abstract
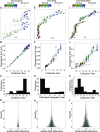
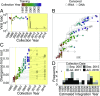
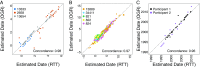
Given that HIV evolution and latent reservoir establishment occur continually within-host, and that latently infected cells can persist long-term, the HIV reservoir should comprise a genetically heterogeneous archive recapitulating within-host HIV evolution. However, this has yet to be conclusively demonstrated, in part due to the challenges of reconstructing within-host reservoir establishment dynamics over long timescales. We developed a phylogenetic framework to reconstruct the integration dates of individual latent HIV lineages. The framework first involves inference and rooting of a maximum-likelihood phylogeny relating plasma HIV RNA sequences serially sampled before the initiation of suppressive antiretroviral therapy, along with putative latent sequences sampled thereafter. A linear model relating root-to-tip distances of plasma HIV RNA sequences to their sampling dates is used to convert root-to-tip distances of putative latent lineages to their establishment (integration) dates. Reconstruction of the ages of putative latent sequences sampled from chronically HIV-infected individuals up to 10 y following initiation of suppressive therapy revealed a genetically heterogeneous reservoir that recapitulated HIV's within-host evolutionary history. Reservoir sequences were interspersed throughout multiple within-host lineages, with the oldest dating to >20 y before sampling; historic genetic bottleneck events were also recorded therein. Notably, plasma HIV RNA sequences isolated from a viremia blip in an individual receiving otherwise suppressive therapy were highly genetically diverse and spanned a 20-y age range, suggestive of spontaneous in vivo HIV reactivation from a large latently infected cell pool. Our framework for reservoir dating provides a potentially powerful addition to the HIV persistence research toolkit.
Keywords: HIV; evolution; latency; phylogenetics; reservoir.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

